
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,715,789 – 13,715,905 |
| Length | 116 |
| Max. P | 0.999826 |

| Location | 13,715,789 – 13,715,905 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.62 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -13.29 |
| Energy contribution | -14.57 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.41 |
| SVM decision value | 4.18 |
| SVM RNA-class probability | 0.999826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
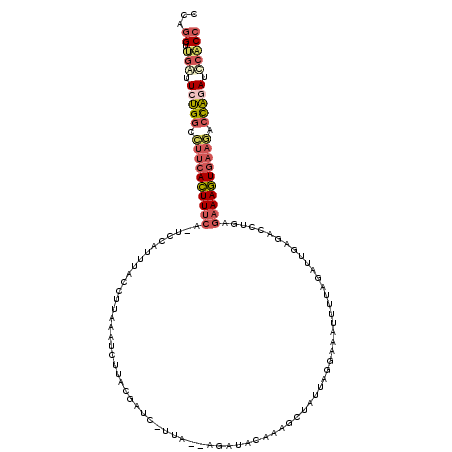
>3R_DroMel_CAF1 13715789 116 + 27905053 CCAGGCUGAUUCUGGCCUUCACUUUCA-UCCAUUUACCUUAAAUCUUACGAUC-UUA--AGAUACAAAGCUAUUAGGAAAUUUACGAGAGAGACCUAAGAAAGUGAAGACUAGAUCCACC ...((.((..(((((.((((((((((.-..............((((((.....-.))--)))).........(((((...(((.(....))))))))))))))))))).)))))..)))) ( -30.50) >DroSec_CAF1 25567 116 + 1 CCAGGUUGAUUCUGGCCUUCACUUUCA-UCCAUUUACCUUAAAUCUUACGAUC-UUA--AGAAACAAAGCUAUUAGGAAGUUUUAGAUUGUGAGCUGGGAAAGUCCCCCCCAAAUUCACC ...(((.((((.(((.....((((((.-.(((............(((((((((-(..--.(((((....((....))..))))))))))))))).))))))))).....))))))).))) ( -22.32) >DroSim_CAF1 29312 116 + 1 CCAGGUUGAUUCUGGCCUUCACUUUCA-UCCAUUUACCUUAAAUCUCACGAUC-UUA--AGUUACAAAGCUAUUAGGAAGUUAUAGAUUGUGAGCUGGGAAAGUGAAGACCAAAUCCACC ...(((.((((.(((.((((((((((.-.(((............(((((((((-(((--(.((.(.((....)).).)).))).)))))))))).))))))))))))).))))))).))) ( -35.12) >DroEre_CAF1 26078 112 + 1 CCAGGUCGGUUCUGGCCUUCACUUUCA-UCAAUUCAGCUUAAAUCGUACGAU-------AGCUACAAAGCUAUUCGGAUACUUCAGAUUGAGACCUGAGAAAGUGAAGACUAGAGCCGCC ...((.(((((((((.((((((((((.-(((......(((((.((...((((-------((((....))))).)))((....)).)))))))...))))))))))))).))))))))))) ( -44.00) >DroAna_CAF1 15216 106 + 1 CCAAGCUGGAUCCGUCUUUCAGUUCAGGACUAUGUGAGUUUAAUAUUAAAACUUUUAAUAUAUUCAAAGUU--UCG------------UAAAAUCUGUAAAACUGAAAGUCGGAUCAUCC ........((((((.(((((((((((((..((((.((.(((.(((((((.....)))))))....))).))--.))------------))...))))...))))))))).)))))).... ( -29.00) >consensus CCAGGUUGAUUCUGGCCUUCACUUUCA_UCCAUUUACCUUAAAUCUUACGAUC_UUA__AGAUACAAAGCUAUUAGGAAAUUUUAGAUUGAGACCUGAGAAAGUGAAGACCAGAUCCACC ...((.(((.(((((.((((((((((........................................................................)))))))))).))))).))))) (-13.29 = -14.57 + 1.28)

| Location | 13,715,789 – 13,715,905 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.62 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.44 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13715789 116 - 27905053 GGUGGAUCUAGUCUUCACUUUCUUAGGUCUCUCUCGUAAAUUUCCUAAUAGCUUUGUAUCU--UAA-GAUCGUAAGAUUUAAGGUAAAUGGA-UGAAAGUGAAGGCCAGAAUCAGCCUGG (((.(((((.(((((((((((((((((................)))))...((...(((((--((.-((((....)))))))))))...)).-.)))))))))))).)).))).)))... ( -38.39) >DroSec_CAF1 25567 116 - 1 GGUGAAUUUGGGGGGGACUUUCCCAGCUCACAAUCUAAAACUUCCUAAUAGCUUUGUUUCU--UAA-GAUCGUAAGAUUUAAGGUAAAUGGA-UGAAAGUGAAGGCCAGAAUCAACCUGG (((((.(((((..(((.....)))...((((..((.......(((.....((((((..(((--((.-.....)))))..))))))....)))-.))..))))...))))).)).)))... ( -28.50) >DroSim_CAF1 29312 116 - 1 GGUGGAUUUGGUCUUCACUUUCCCAGCUCACAAUCUAUAACUUCCUAAUAGCUUUGUAACU--UAA-GAUCGUGAGAUUUAAGGUAAAUGGA-UGAAAGUGAAGGCCAGAAUCAACCUGG (((.(((((((((((((((((((((....((((.((((.........))))..)))).(((--(.(-((((....))))).))))...))).-.))))))))))))))).))).)))... ( -42.10) >DroEre_CAF1 26078 112 - 1 GGCGGCUCUAGUCUUCACUUUCUCAGGUCUCAAUCUGAAGUAUCCGAAUAGCUUUGUAGCU-------AUCGUACGAUUUAAGCUGAAUUGA-UGAAAGUGAAGGCCAGAACCGACCUGG (((((.(((.((((((((((((((((((....))))))......(((.(((((....))))-------))))..((((((.....)))))).-.)))))))))))).))).))).))... ( -42.40) >DroAna_CAF1 15216 106 - 1 GGAUGAUCCGACUUUCAGUUUUACAGAUUUUA------------CGA--AACUUUGAAUAUAUUAAAAGUUUUAAUAUUAAACUCACAUAGUCCUGAACUGAAAGACGGAUCCAGCUUGG ....((((((.(((((((((...(((......------------.((--((((((..........))))))))........(((.....))).)))))))))))).))))))........ ( -25.70) >consensus GGUGGAUCUGGUCUUCACUUUCCCAGCUCUCAAUCUAAAACUUCCUAAUAGCUUUGUAUCU__UAA_GAUCGUAAGAUUUAAGGUAAAUGGA_UGAAAGUGAAGGCCAGAAUCAACCUGG (((((.((((((((((((((((...(....)....................................((((....))))...............)))))))))))))))).))).))... (-21.48 = -21.44 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:30 2006