
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
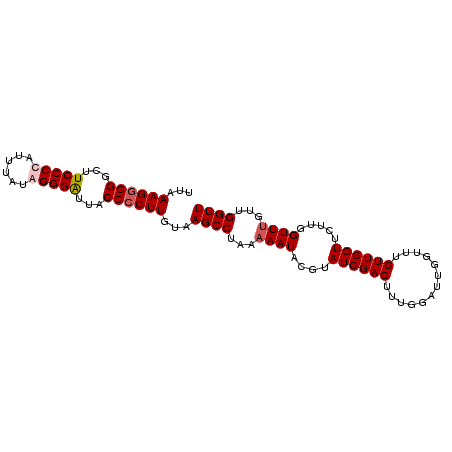
| Location | 13,706,235 – 13,706,331 |
| Length | 96 |
| Max. P | 0.974746 |

| Location | 13,706,235 – 13,706,331 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13706235 96 + 27905053 UUGAAGGGGGCUUCCCCAUUUAUAGGGGUUACCCCUUGUAAGCCUAAAAAUACGUAUCGACUUUGGAUUGGUUUGUUGGUUCUUGGUUUAUUGGUU ...((((((....((((.......))))...))))))(((((((.......(((.((((((....).))))).)))........)))))))..... ( -27.26) >DroSec_CAF1 16004 95 + 1 UUAAAGGGGGCUUCCCCUUUUAUGGGGAUUACCCCUUGUAAGCCUAA-AAUACGUAUCGACUUUGGAUUGGUUUGUUGGUUCUUGGUUUGUUGGUU ...((((((...(((((......)))))...))))))...((((..(-(((....((((((.............)))))).....))))...)))) ( -30.12) >DroSim_CAF1 16667 96 + 1 UUAAAGGGGGCUUCCCCUUUUAUAGGGAUUACCCCUUGUAAGCCUAAAAAUACGUAUCGACUUUGGAUUGGUUUGUUGGUUCUUGGUUUGUUGGUU ...((((((...((((........))))...))))))...((((...((((....((((((.............)))))).....))))...)))) ( -25.92) >DroYak_CAF1 24789 96 + 1 UUAAAGUGGGUUUCCCCAUUAUUGGGGGAUACCCCUUGUAAGCCUAAAAAUACGUAUCGACUUUGGAUUGGAUUGUUGGUUCUUGGUUUGCUGGUU ...(((.(((((((((((....))))))).)))))))(((((((.........((....))...((((..(....)..))))..)))))))..... ( -29.50) >consensus UUAAAGGGGGCUUCCCCAUUUAUAGGGAUUACCCCUUGUAAGCCUAAAAAUACGUAUCGACUUUGGAUUGGUUUGUUGGUUCUUGGUUUGUUGGUU ...((((((...(((((......)))))...))))))...((((...((((....((((((.............)))))).....))))...)))) (-24.17 = -24.92 + 0.75)


| Location | 13,706,235 – 13,706,331 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -17.20 |
| Energy contribution | -17.02 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13706235 96 - 27905053 AACCAAUAAACCAAGAACCAACAAACCAAUCCAAAGUCGAUACGUAUUUUUAGGCUUACAAGGGGUAACCCCUAUAAAUGGGGAAGCCCCCUUCAA ..............(((................(((((..............)))))....(((((..((((.......))))..))))).))).. ( -20.84) >DroSec_CAF1 16004 95 - 1 AACCAACAAACCAAGAACCAACAAACCAAUCCAAAGUCGAUACGUAUU-UUAGGCUUACAAGGGGUAAUCCCCAUAAAAGGGGAAGCCCCCUUUAA .................................(((((..........-...)))))..((((((...(((((......)))))...))))))... ( -21.42) >DroSim_CAF1 16667 96 - 1 AACCAACAAACCAAGAACCAACAAACCAAUCCAAAGUCGAUACGUAUUUUUAGGCUUACAAGGGGUAAUCCCUAUAAAAGGGGAAGCCCCCUUUAA .................................(((((..............)))))..((((((...(((((......)))))...))))))... ( -19.14) >DroYak_CAF1 24789 96 - 1 AACCAGCAAACCAAGAACCAACAAUCCAAUCCAAAGUCGAUACGUAUUUUUAGGCUUACAAGGGGUAUCCCCCAAUAAUGGGGAAACCCACUUUAA .................................(((((..............)))))..(((((((.(((((.......))))).)))).)))... ( -19.54) >consensus AACCAACAAACCAAGAACCAACAAACCAAUCCAAAGUCGAUACGUAUUUUUAGGCUUACAAGGGGUAACCCCCAUAAAAGGGGAAGCCCCCUUUAA .................................(((((..............)))))....(((((..((((.......))))..)))))...... (-17.20 = -17.02 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:23 2006