
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,696,319 – 13,696,439 |
| Length | 120 |
| Max. P | 0.959967 |

| Location | 13,696,319 – 13,696,439 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -48.56 |
| Consensus MFE | -45.30 |
| Energy contribution | -45.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
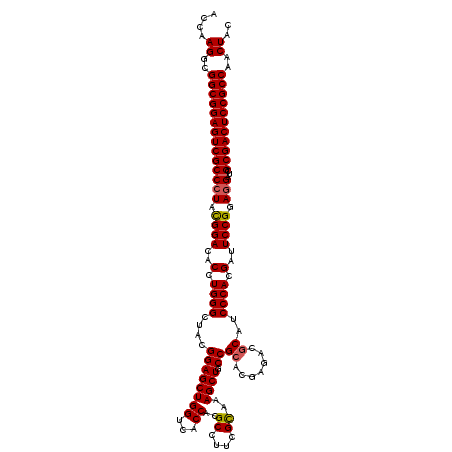
>3R_DroMel_CAF1 13696319 120 + 27905053 ACCAAGGCGGCGGAGUCGCCGUACGGACACCUGGGCUACGGAGCUGGUCACCACGCCUUCGCAAAGCUGCCGCACGAGACGCAUCCCACGAUUCCGGAGGAUCGCGACUCCGCCAACUAC ....((..((((((((((((((.(((.((.((..((...((.(((((...))).))))..))..)).))))).)))....(.((((..((....))..))))))))))))))))..)).. ( -49.40) >DroSec_CAF1 6147 120 + 1 ACCAAGGCGGCGGAGUCGCCCUAUGGACACCUGGGCUACGGAGCUGGUCACCACGCCUUCGCAAAGCUGCCGCACGAGACGCAUCCCACGAUUCCGGAGGAUCGCGACUCCGCCAACUAC ....((..(((((((((((((...((...)).))))..(((((((((...))..((....))..)))).)))........((((((..((....))..)))).)))))))))))..)).. ( -47.00) >DroSim_CAF1 6214 120 + 1 ACCAAGGCGGCGGAGUCGCCCUAUGGACACCUGGGCUACGGAGCUGGUCACCACGCCUUCGCAAAGCUGCCGCACGAGACGCAUCCCACGAUUCCGGAGGAUCGCGACUCCGCCAACUAC ....((..(((((((((((((...((...)).))))..(((((((((...))..((....))..)))).)))........((((((..((....))..)))).)))))))))))..)).. ( -47.00) >DroEre_CAF1 6489 120 + 1 ACCAAGGCGGCGGAGUCGCCCUACGGACACCUGGGCUACGGAGCUGGUCACCACGCCUUCGCAAAGCUGCCGCUCGAGACGCAUCCCACGAUUCCGGAGGAUCGCGACUCCGCCAACUAC ....((..((((((((((((((.((((..(.((((...((((((.(((......)))...(((....))).))))....))...)))).)..)))).)))...)))))))))))..)).. ( -49.00) >DroYak_CAF1 6840 120 + 1 ACCAAGGCGGCGGAGUCGCCCUACGGACACUUGGGCUACGGAGCUGGUCACCACGCCUUCGUGAAGCUGCCGCACGAGACUCAUCCCACGAUUCCGGAGGAUCGCGACUCCGCCAACUAC ....((..((((((((((((((.((((..(.((((.....(((((.((...((((....))))..((....)))).)).)))..)))).)..)))).)))...)))))))))))..)).. ( -50.40) >consensus ACCAAGGCGGCGGAGUCGCCCUACGGACACCUGGGCUACGGAGCUGGUCACCACGCCUUCGCAAAGCUGCCGCACGAGACGCAUCCCACGAUUCCGGAGGAUCGCGACUCCGCCAACUAC ....((..((((((((((((((.((((..(.((((....((((((((...))..((....))..)))).))((.......))..)))).)..)))).)))...)))))))))))..)).. (-45.30 = -45.30 + 0.00)

| Location | 13,696,319 – 13,696,439 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -57.54 |
| Consensus MFE | -53.40 |
| Energy contribution | -54.24 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13696319 120 - 27905053 GUAGUUGGCGGAGUCGCGAUCCUCCGGAAUCGUGGGAUGCGUCUCGUGCGGCAGCUUUGCGAAGGCGUGGUGACCAGCUCCGUAGCCCAGGUGUCCGUACGGCGACUCCGCCGCCUUGGU ..((.((((((((((((.((((..((....))..))))......(((((((((.((..((...((.(((....)..)).))...))..)).)).))))))))))))))))))).)).... ( -57.40) >DroSec_CAF1 6147 120 - 1 GUAGUUGGCGGAGUCGCGAUCCUCCGGAAUCGUGGGAUGCGUCUCGUGCGGCAGCUUUGCGAAGGCGUGGUGACCAGCUCCGUAGCCCAGGUGUCCAUAGGGCGACUCCGCCGCCUUGGU ..((.((((((((((((...(((..(((..(.((((.((((....(.((((..(((.(((....))).)))..)).)).))))).)))).)..)))..))))))))))))))).)).... ( -55.50) >DroSim_CAF1 6214 120 - 1 GUAGUUGGCGGAGUCGCGAUCCUCCGGAAUCGUGGGAUGCGUCUCGUGCGGCAGCUUUGCGAAGGCGUGGUGACCAGCUCCGUAGCCCAGGUGUCCAUAGGGCGACUCCGCCGCCUUGGU ..((.((((((((((((...(((..(((..(.((((.((((....(.((((..(((.(((....))).)))..)).)).))))).)))).)..)))..))))))))))))))).)).... ( -55.50) >DroEre_CAF1 6489 120 - 1 GUAGUUGGCGGAGUCGCGAUCCUCCGGAAUCGUGGGAUGCGUCUCGAGCGGCAGCUUUGCGAAGGCGUGGUGACCAGCUCCGUAGCCCAGGUGUCCGUAGGGCGACUCCGCCGCCUUGGU ..((.((((((((((((...(((.((((..(.((((.((((....((((((..(((.(((....))).)))..)).)))))))).)))).)..)))).))))))))))))))).)).... ( -63.10) >DroYak_CAF1 6840 120 - 1 GUAGUUGGCGGAGUCGCGAUCCUCCGGAAUCGUGGGAUGAGUCUCGUGCGGCAGCUUCACGAAGGCGUGGUGACCAGCUCCGUAGCCCAAGUGUCCGUAGGGCGACUCCGCCGCCUUGGU ..((.((((((((((((...(((.((((....(((((((.....)))((((.(((((((((....))))).....))))))))..))))....)))).))))))))))))))).)).... ( -56.20) >consensus GUAGUUGGCGGAGUCGCGAUCCUCCGGAAUCGUGGGAUGCGUCUCGUGCGGCAGCUUUGCGAAGGCGUGGUGACCAGCUCCGUAGCCCAGGUGUCCGUAGGGCGACUCCGCCGCCUUGGU ..((.((((((((((((...(((.((((..(.((((.((((....(.((((..(((.(((....))).)))..)).)).))))).)))).)..)))).))))))))))))))).)).... (-53.40 = -54.24 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:13 2006