
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,695,188 – 13,695,401 |
| Length | 213 |
| Max. P | 0.999724 |

| Location | 13,695,188 – 13,695,297 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 98.62 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13695188 109 - 27905053 AUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUACACGCCCCUACAUGACGACGCAUCGCCUCCCUGCAAUCCCACAUGCAAAUCACCCAAACAAGAGCACAAA ......((((((((((..(((.(((((((((........)))))))))....)).)..)))..))))...((((........))))..............)))...... ( -29.70) >DroSec_CAF1 5041 109 - 1 AUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUACACGCCCCUACAUGACGACGCAUCGCCUCCCUGCAAUCCCACAUGCAAAUCACCCAAACAAGAGCACAAA ......((((((((((..(((.(((((((((........)))))))))....)).)..)))..))))...((((........))))..............)))...... ( -29.70) >DroSim_CAF1 5085 109 - 1 AUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUACACGCCCCUACAUGACGACGCAUCGCCUCCCUGCAAUCCCACAUGCAAAUCACCCAAACAAGAGCACAAA ......((((((((((..(((.(((((((((........)))))))))....)).)..)))..))))...((((........))))..............)))...... ( -29.70) >DroEre_CAF1 5358 109 - 1 AUAAAUCUCAGCCUGCUGGUCCAGGGGCGUGACAUGCUACACGCCCCUACACGACGACGCAUCGCCUCCCUGCAAUCCCACAUGCAAAUCACCCAAACAAGAGCACAAA ......(((.((.((((.(((.(((((((((........)))))))))....))).).)))..)).....((((........))))..............)))...... ( -29.70) >consensus AUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUACACGCCCCUACAUGACGACGCAUCGCCUCCCUGCAAUCCCACAUGCAAAUCACCCAAACAAGAGCACAAA ......((((((((((...((.(((((((((........)))))))))....))....)))..))))...((((........))))..............)))...... (-26.58 = -26.82 + 0.25)

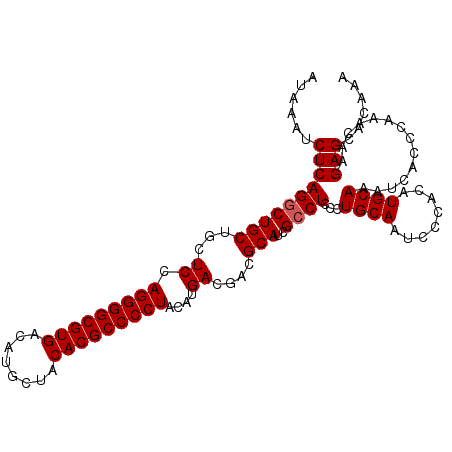
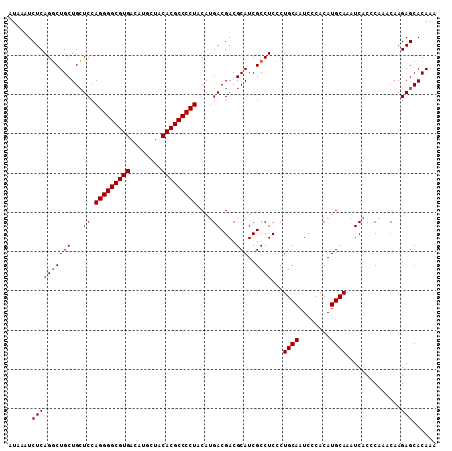
| Location | 13,695,217 – 13,695,337 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -47.50 |
| Consensus MFE | -41.53 |
| Energy contribution | -42.06 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13695217 120 + 27905053 UGGGAUUGCAGGGAGGCGAUGCGUCGUCAUGUAGGGGCGUGUAGCAUGUCACGCCCCUGGAGCAGCAGCCUGAGAUUUAUGUUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCA (((.(((.((((..(((((....))))).(.((((((((((........)))))))))).).......)))).((((((....(((((((....)))))))))))))))).)))...... ( -48.00) >DroSec_CAF1 5070 120 + 1 UGGGAUUGCAGGGAGGCGAUGCGUCGUCAUGUAGGGGCGUGUAGCAUGUCACGCCCCUGGAGCAGCAGCCUGAGAUUUAUGCUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCA (((.(((.((((..(((((....))))).(.((((((((((........)))))))))).).......)))).((((((((((..((......))..))))))))))))).)))...... ( -48.90) >DroSim_CAF1 5114 120 + 1 UGGGAUUGCAGGGAGGCGAUGCGUCGUCAUGUAGGGGCGUGUAGCAUGUCACGCCCCUGGAGCAGCAGCCUGAGAUUUAUGUUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCA (((.(((.((((..(((((....))))).(.((((((((((........)))))))))).).......)))).((((((....(((((((....)))))))))))))))).)))...... ( -48.00) >DroEre_CAF1 5387 120 + 1 UGGGAUUGCAGGGAGGCGAUGCGUCGUCGUGUAGGGGCGUGUAGCAUGUCACGCCCCUGGACCAGCAGGCUGAGAUUUAUGUUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCA .((...(((.....(((((....)))))((.((((((((((........)))))))))).))..)))(((...((((((....(((((((....)))))))))))))...)))....)). ( -51.00) >DroYak_CAF1 5804 101 + 1 UGAAACUGCA-------------------UGUAGGGGCGUGUAGCAUGUCACGCCCCUAGAGCAGCAGCCUGAGAUUUAUGUUUGCUUUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCA .....((((.-------------------(.((((((((((........)))))))))).)))))..((((..((((((((((..(((....)))..)))))))))).......)))).. ( -41.60) >consensus UGGGAUUGCAGGGAGGCGAUGCGUCGUCAUGUAGGGGCGUGUAGCAUGUCACGCCCCUGGAGCAGCAGCCUGAGAUUUAUGUUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCA .((..((((.....(((((....))))).(.((((((((((........)))))))))).)...))))((((..(((.((...(((((((....)))))))...)).)))..)))).)). (-41.53 = -42.06 + 0.53)


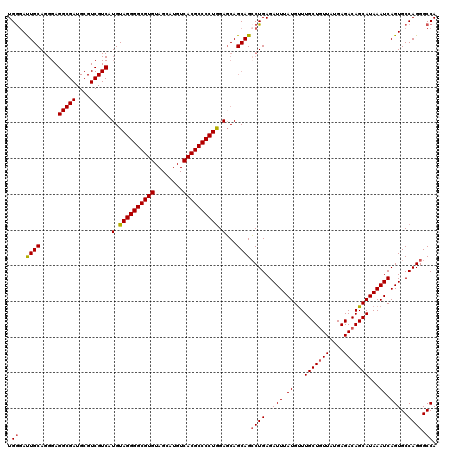
| Location | 13,695,217 – 13,695,337 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -32.76 |
| Energy contribution | -33.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13695217 120 - 27905053 UGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAACAUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUACACGCCCCUACAUGACGACGCAUCGCCUCCCUGCAAUCCCA .((.....(((..((((((((((((......))))))....))))))..(((((((..(((.(((((((((........)))))))))....)).)..)))..))))...)))....)). ( -38.80) >DroSec_CAF1 5070 120 - 1 UGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAGCAUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUACACGCCCCUACAUGACGACGCAUCGCCUCCCUGCAAUCCCA .((.....(((..((((((((((..((......))..))))))))))..(((((((..(((.(((((((((........)))))))))....)).)..)))..))))...)))....)). ( -40.30) >DroSim_CAF1 5114 120 - 1 UGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAACAUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUACACGCCCCUACAUGACGACGCAUCGCCUCCCUGCAAUCCCA .((.....(((..((((((((((((......))))))....))))))..(((((((..(((.(((((((((........)))))))))....)).)..)))..))))...)))....)). ( -38.80) >DroEre_CAF1 5387 120 - 1 UGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAACAUAAAUCUCAGCCUGCUGGUCCAGGGGCGUGACAUGCUACACGCCCCUACACGACGACGCAUCGCCUCCCUGCAAUCCCA .(((...(((...((((((((((((......))))))....))))))...)))((((.(((.(((((((((........)))))))))....))).).)))..))).............. ( -42.90) >DroYak_CAF1 5804 101 - 1 UGGCCCUGGCACUGAUUUAUGCUGUCUCAUAAAAGCAAACAUAAAUCUCAGGCUGCUGCUCUAGGGGCGUGACAUGCUACACGCCCCUACA-------------------UGCAGUUUCA .((((.(((....((((((((.((.((......))))..)))))))))))))))(((((..((((((((((........))))))))))..-------------------.))))).... ( -38.80) >consensus UGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAACAUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUACACGCCCCUACAUGACGACGCAUCGCCUCCCUGCAAUCCCA .((((((((....((((((((((((......))))))....)))))))))))..))).....(((((((((........)))))))))................................ (-32.76 = -33.16 + 0.40)



| Location | 13,695,257 – 13,695,370 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -31.90 |
| Energy contribution | -31.46 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13695257 113 - 27905053 -------GUCCCCAAUCCGCAGGCCAUGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAACAUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUAC -------((((((...((((((((.(((....))))))))))).(((((....((((((((((((......))))))....))))))...(((....)))))))))).).)))....... ( -38.50) >DroSec_CAF1 5110 114 - 1 ------GAUCCCCAGUCCCCAGGCCAUGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAGCAUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUAC ------......((((.((..(((((((..(....)..)))))))..)).)))).....((((((......))))))(((((.....(((.(((.(((...))).))).))).))))).. ( -38.20) >DroSim_CAF1 5154 114 - 1 ------GAUCCCCAGUCCCCAGGCCAUGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAACAUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUAC ------.....((((..(((((((.(((....)))))))).))..))))....((((((((((((......))))))....))))))(((.(((.(((...))).))).)))........ ( -36.60) >DroEre_CAF1 5427 120 - 1 UUCCCACAUUCCCAGUCCCCAGGCCAUGUCCCCGUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAACAUAAAUCUCAGCCUGCUGGUCCAGGGGCGUGACAUGCUAC .....................(((.(((((..(((.((((.((((..(((...((((((((((((......))))))....))))))...)))....)))))))).))).)))))))).. ( -39.50) >DroYak_CAF1 5825 103 - 1 -----------------CCCAGACCAAGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAAAAGCAAACAUAAAUCUCAGGCUGCUGCUCUAGGGGCGUGACAUGCUAC -----------------...............((((((...(.((((((....((((((((.((.((......))))..))))))))...(((....))))))))).)..)))))).... ( -27.10) >consensus _______AUCCCCAGUCCCCAGGCCAUGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAACAUAAAUCUCAGGCUGCUGCUCCAGGGGCGUGACAUGCUAC ................................((((((...(.((((((....((((((((((((......))))))....))))))...(((....))))))))).)..)))))).... (-31.90 = -31.46 + -0.44)



| Location | 13,695,297 – 13,695,401 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.96 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -29.87 |
| Energy contribution | -32.05 |
| Covariance contribution | 2.18 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13695297 104 + 27905053 GUUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCACAGACAUGGGGACAUGGCCUGCGGAUUGGGGAC----------------CUGGUCUGGGGGACAGUGGCAGCGACACUGU ...(((((((....))))))).....(((((.(...(((((...((((....)))).(((.(((((..(....----------------)..))))).)))...)))))...).))))). ( -47.30) >DroSec_CAF1 5150 106 + 1 GCUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCACAGACAUGGGGACAUGGCCUGGGGACUGGGGAUC--------------UCUGGUCUGGGGGACAGUGGCAGCGACACUGU ...(((((((....))))))).....(((((.(...(((((...((((....)))).(((..((((..((....--------------))..))))..)))...)))))...).))))). ( -44.60) >DroSim_CAF1 5194 106 + 1 GUUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCACAGACAUGGGGACAUGGCCUGGGGACUGGGGAUC--------------UCUGGUCUGGGGGACAGUGGCAGCGACACUGU ...(((((((....))))))).....(((((.(...(((((...((((....)))).(((..((((..((....--------------))..))))..)))...)))))...).))))). ( -44.60) >DroEre_CAF1 5467 120 + 1 GUUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCACAGACACGGGGACAUGGCCUGGGGACUGGGAAUGUGGGAAUGUGGGGAUCUGGUCUGGGGGACAGCGGCAGCGACACUGU ...(((((((.(.((((.((((......))))((((.(((((..((((....)....((.(....).))....)))....)))))...)))))))).)..)))))))((((.....)))) ( -41.00) >DroYak_CAF1 5865 83 + 1 GUUUGCUUUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCACAGACAUGGGGACUUGGUCUGGG-------------------------------------CACAUCGGCAGCGACACUGU ((((((.((((((......))))))....((((..((((.(((((..(....)...)))))))-------------------------------------).)...)))))))).))... ( -27.80) >consensus GUUUGCUGUUAUGAGACAGCAUAAAUCAGUGCCAGGGCCACAGACAUGGGGACAUGGCCUGGGGACUGGGGAU_______________UCUGGUCUGGGGGACAGUGGCAGCGACACUGU ...(((((((....))))))).....(((((.(...(((((...((((....)))).(((..(((((((....................)))))))..)))...)))))...).))))). (-29.87 = -32.05 + 2.18)


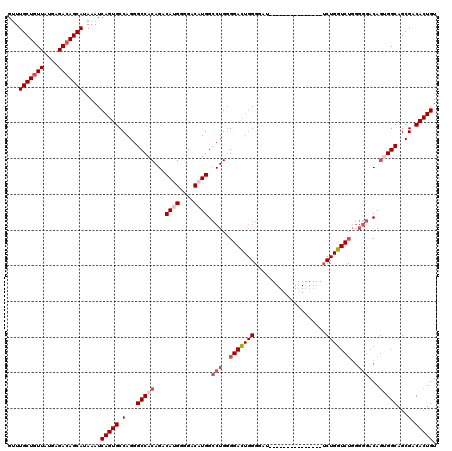
| Location | 13,695,297 – 13,695,401 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.96 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13695297 104 - 27905053 ACAGUGUCGCUGCCACUGUCCCCCAGACCAG----------------GUCCCCAAUCCGCAGGCCAUGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAAC .((((((((..(((.(((((.....)).)))----------------...........((((((.(((....))))))))))))..)))))))).....((((((......))))))... ( -35.10) >DroSec_CAF1 5150 106 - 1 ACAGUGUCGCUGCCACUGUCCCCCAGACCAGA--------------GAUCCCCAGUCCCCAGGCCAUGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAGC .(((((((.(((...(((.....)))..))).--------------))...((((..(((((((.(((....)))))))).))..))))))))).....((((((......))))))... ( -34.00) >DroSim_CAF1 5194 106 - 1 ACAGUGUCGCUGCCACUGUCCCCCAGACCAGA--------------GAUCCCCAGUCCCCAGGCCAUGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAAC .(((((((.(((...(((.....)))..))).--------------))...((((..(((((((.(((....)))))))).))..))))))))).....((((((......))))))... ( -34.00) >DroEre_CAF1 5467 120 - 1 ACAGUGUCGCUGCCGCUGUCCCCCAGACCAGAUCCCCACAUUCCCACAUUCCCAGUCCCCAGGCCAUGUCCCCGUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAAC .((((((((..(((((.(((.....))).........((((....((((..((........))..))))....))))..)))))..)))))))).....((((((......))))))... ( -33.60) >DroYak_CAF1 5865 83 - 1 ACAGUGUCGCUGCCGAUGUG-------------------------------------CCCAGACCAAGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAAAAGCAAAC .(((((((((..(.((((((-------------------------------------....(((...)))..)))))).)..))...))))))).....((((..........))))... ( -24.30) >consensus ACAGUGUCGCUGCCACUGUCCCCCAGACCAGA_______________AUCCCCAGUCCCCAGGCCAUGUCCCCAUGUCUGUGGCCCUGGCACUGAUUUAUGCUGUCUCAUAACAGCAAAC .((((((((..(((((((.....))).................................(((((.(((....))))))))))))..)))))))).....((((((......))))))... (-28.90 = -29.38 + 0.48)

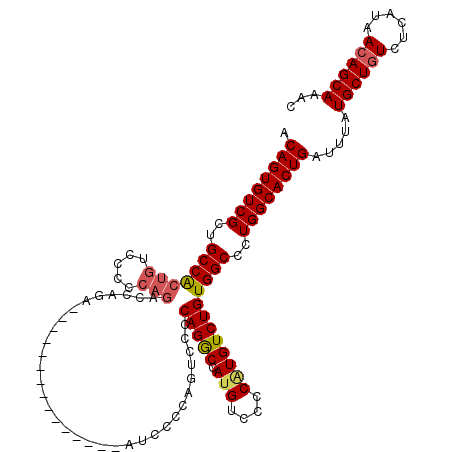

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:06 2006