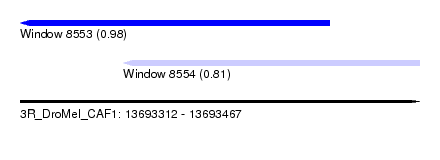
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,693,312 – 13,693,467 |
| Length | 155 |
| Max. P | 0.977206 |

| Location | 13,693,312 – 13,693,432 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -26.78 |
| Energy contribution | -26.66 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13693312 120 - 27905053 AAUUAGGUGUGGCAAAACGCUGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGACGUGCAGAGAAAACACACCGAUAAAGUACAAAAUGAAUGA .....((((((........((((((..............((((.((((........)))).)))).((((......)))).))))))......))))))..................... ( -31.74) >DroSec_CAF1 3208 120 - 1 AAUUAGGUGUGGCAAAACGCUGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGACGUGCAGAGGAAACACACCGAUAAAGUACAAAAUGAAUGA .....((((((......(.((((((..............((((.((((........)))).)))).((((......)))).)))))).)....))))))..................... ( -32.20) >DroSim_CAF1 3222 120 - 1 AAUUAGGUGUGGCAAAACGCUGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGACGUGCAGAGGAAACACACCGAUAAAGUACAAAAUGAAUGA .....((((((......(.((((((..............((((.((((........)))).)))).((((......)))).)))))).)....))))))..................... ( -32.20) >DroEre_CAF1 3213 120 - 1 AAUUUGGUGUGGCAAAACGCACCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGACAAUGGGACGUGCAGAGGAAACACACCAAUAAAGUGCAAAAUGAAUGA ...((((((((.(.....(((((((((.....)))))..((((.((((........)))).)))).((((......)))).))))....)...))))))))................... ( -31.00) >DroYak_CAF1 3266 120 - 1 AAUUAUGUGAGGCAAAACGCAGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGACGUGCAGAGGAAACACACCGAUAAAGUACAAAAUCAAUGA ......(((........(.((((((((.....)))))))).)......((((((.......((.(.((((......)))).).))...)))))).)))...................... ( -25.10) >consensus AAUUAGGUGUGGCAAAACGCUGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGACGUGCAGAGGAAACACACCGAUAAAGUACAAAAUGAAUGA .....((((..((.....))..))))..(((((..((((((.......((((((.......((.(.((((......)))).).))...))))))...)))))).))).)).......... (-26.78 = -26.66 + -0.12)


| Location | 13,693,352 – 13,693,467 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -30.31 |
| Consensus MFE | -25.96 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13693352 115 - 27905053 AAACUGA-----AGAGUACAGUGCUGGUAAAUGUGGGGUAAAUUAGGUGUGGCAAAACGCUGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGA (((((((-----...(..(((..(((((.............(((((.((..((.....))..))))))).)))))..)))..)..)))))))..............((((......)))) ( -29.44) >DroSec_CAF1 3248 115 - 1 AAACUGA-----AGAGUACAGUGCUGGUAAAUGUGGGGUAAAUUAGGUGUGGCAAAACGCUGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGA (((((((-----...(..(((..(((((.............(((((.((..((.....))..))))))).)))))..)))..)..)))))))..............((((......)))) ( -29.44) >DroSim_CAF1 3262 115 - 1 AAACUGA-----AGAGUACAGUGCUGGUAAAUGUGGGGUAAAUUAGGUGUGGCAAAACGCUGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGA (((((((-----...(..(((..(((((.............(((((.((..((.....))..))))))).)))))..)))..)..)))))))..............((((......)))) ( -29.44) >DroEre_CAF1 3253 120 - 1 AAACUAAAACUGAGAGUACAGUGCUGGUAAAUGUGGGGUAAAUUUGGUGUGGCAAAACGCACCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGACAAUGGGA .....((((((((..(..(((..(((((.....(((........(((((((......))))))))))...)))))..)))..)..)))))))).............((((......)))) ( -34.20) >DroYak_CAF1 3306 120 - 1 AAACUAAAACUGAGAGUACAGUGCUGGUAAAUGUGGGGUAAAUUAUGUGAGGCAAAACGCAGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGA .....((((((((..(..(((..(((((.............((((.(((..((.....))..))))))).)))))..)))..)..)))))))).............((((......)))) ( -29.04) >consensus AAACUGA_____AGAGUACAGUGCUGGUAAAUGUGGGGUAAAUUAGGUGUGGCAAAACGCUGCACUAAUGACUAGUGUUGGGCAAUUAGUUUUCAAUAAUGGCUCUUCCCGAAAAUGGGA ............(((((.(((..(((((.............((((.(((..((.....))..))))))).)))))..)))....((((........)))).)))))((((......)))) (-25.96 = -26.16 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:59 2006