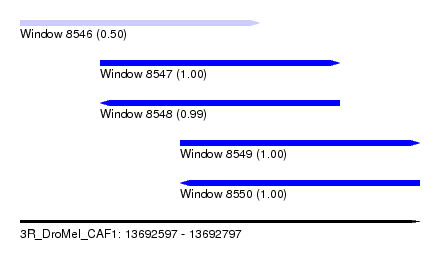
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,692,597 – 13,692,797 |
| Length | 200 |
| Max. P | 0.999892 |

| Location | 13,692,597 – 13,692,717 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -40.64 |
| Energy contribution | -40.88 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13692597 120 + 27905053 UUGCAGCUCUUGUCGGUGUGACACACCUGCUGCUAUGGAUUGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCAUUGGACAGCACUUACGGUGGACUGC ..((((((..((((((((.....)))).((((((...(((.((((...(((...))))))).))).))))))...)))).....)))))).(((((((...........))))))).... ( -42.60) >DroSec_CAF1 2498 120 + 1 UUGCAGCUCUGGUCGGAGUGACACACCUGCUGCUAUGGAUUGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUAC ....((.((((.((((((((...((.(.((((((...(((.((((...(((...))))))).))).))))))).)).)))))...((((((......)).)))).....))))))))).. ( -39.00) >DroSim_CAF1 2513 120 + 1 UUGCAGCUCUUGUCGGAGUGACACACCUGCUGCUAUGGAUUGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUGC ..((((.(((.(((((((((...((.(.((((((...(((.((((...(((...))))))).))).))))))).)).)))))...((((((......)).)))).....))))))))))) ( -44.00) >DroEre_CAF1 2519 120 + 1 UUGCAGCUCUUGUCGGAGUGACACACCUGCUGCUAUGGAUUGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCCCCGCUGGACAGCACUUACGGCGGACUGC ..((((.(((.(((((((((...((.(.((((((...(((.((((...(((...))))))).))).))))))).)).)))))...((((((......)).)))).....))))))))))) ( -42.80) >DroYak_CAF1 2561 120 + 1 UUGCAGCUCUUGUCGGAGUGACACACCUGCUGCUAUGGAUUGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCACUGGACAGCACUUACGGCGGACUGC ..((((.(((.(((((((((...((.(.((((((...(((.((((...(((...))))))).))).))))))).)).)))))...((((((......)).)))).....))))))))))) ( -44.00) >consensus UUGCAGCUCUUGUCGGAGUGACACACCUGCUGCUAUGGAUUGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUGC ..((((.(((.(((((((((...((.(.((((((...(((.((((...(((...))))))).))).))))))).)).)))))...((((((......)).)))).....))))))))))) (-40.64 = -40.88 + 0.24)

| Location | 13,692,637 – 13,692,757 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.92 |
| Mean single sequence MFE | -49.36 |
| Consensus MFE | -47.62 |
| Energy contribution | -46.94 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13692637 120 + 27905053 UGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCAUUGGACAGCACUUACGGUGGACUGCUGGGAAAGUCCAGUGCCCAGCCGCUCCUGAUGUCCAGUGG .(((....)))....((((..(((((((((((...(((((.....((((((......)).))))........((((((........)))))))))))..))))))..)))))..))).). ( -49.10) >DroSec_CAF1 2538 120 + 1 UGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUACUGGGAAAGUCCAGUGCCCAGCCGCUCCUGAUGUCCAGUGG .(((....)))....((((..(((((((((((...((((((....((((((......)).)))).........(((((........)))))))))))..))))))..)))))..))).). ( -48.20) >DroSim_CAF1 2553 120 + 1 UGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUGCUGGGAAAGUCCAGUGCCCAGCCGCUCCUGAUGUCCAGUGG .(((....)))....((((..(((((((((((...((((((....((((((......)).)))).........(((((........)))))))))))..))))))..)))))..))).). ( -49.10) >DroEre_CAF1 2559 120 + 1 UGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCCCCGCUGGACAGCACUUACGGCGGACUGCUGGGCAAGUCCAGCGCCCAGCCGCUCCUGAUGUCCAGUGG ........(((.((((.((.....)).)))).)))((((........)))).((((((((((.((....((((((..(((((((....))))))).)).))))....)).)))))))))) ( -51.90) >DroYak_CAF1 2601 120 + 1 UGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCACUGGACAGCACUUACGGCGGACUGCUGGGGAAGUCAAGUGCCCAGCCGCUCCUGAUGUCCAGUGG .(((....)))....((((..(((((((((((...(((((((...((((((......)).))))((((.((((.....))))...)))).)))))))..))))))..)))))..))).). ( -48.50) >consensus UGUCAUCUGGCUCUGCCUGACCAUCAAGCGGCGCUGGCACUUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUGCUGGGAAAGUCCAGUGCCCAGCCGCUCCUGAUGUCCAGUGG ........(((.((((.((.....)).)))).)))((((........)))).((((((((((.((....((((((.(.((((((....))))))).)).))))....)).)))))))))) (-47.62 = -46.94 + -0.68)


| Location | 13,692,637 – 13,692,757 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.92 |
| Mean single sequence MFE | -50.08 |
| Consensus MFE | -47.08 |
| Energy contribution | -47.12 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13692637 120 - 27905053 CCACUGGACAUCAGGAGCGGCUGGGCACUGGACUUUCCCAGCAGUCCACCGUAAGUGCUGUCCAAUGGAGGCAGCUUAAAGUGCCAGCGCCGCUUGAUGGUCAGGCAGAGCCAGAUGACA ...(((..(((((..((((((..(((((((((((........))))).........(((((((....).))))))....))))))...)))))))))))..)))................ ( -48.10) >DroSec_CAF1 2538 120 - 1 CCACUGGACAUCAGGAGCGGCUGGGCACUGGACUUUCCCAGUAGUCCGCCGUAAGUGCUGUCCAAUGGAGGCAGCUUAAAGUGCCAGCGCCGCUUGAUGGUCAGGCAGAGCCAGAUGACA (((.((((((.((...(((((.(((((((((......))))).)))))))))...)).)))))).))).(((........(((....))).(((((.....)))))...)))........ ( -50.40) >DroSim_CAF1 2553 120 - 1 CCACUGGACAUCAGGAGCGGCUGGGCACUGGACUUUCCCAGCAGUCCGCCGUAAGUGCUGUCCAAUGGAGGCAGCUUAAAGUGCCAGCGCCGCUUGAUGGUCAGGCAGAGCCAGAUGACA ...(((..(((((..((((((..(((((((((((........))))).........(((((((....).))))))....))))))...)))))))))))..)))................ ( -48.10) >DroEre_CAF1 2559 120 - 1 CCACUGGACAUCAGGAGCGGCUGGGCGCUGGACUUGCCCAGCAGUCCGCCGUAAGUGCUGUCCAGCGGGGGCAGCUUAAAGUGCCAGCGCCGCUUGAUGGUCAGGCAGAGCCAGAUGACA ((.(((((((.((...(((((.(((((((((......))))).)))))))))...)).))))))).)).(((........(((....))).(((((.....)))))...)))........ ( -54.00) >DroYak_CAF1 2601 120 - 1 CCACUGGACAUCAGGAGCGGCUGGGCACUUGACUUCCCCAGCAGUCCGCCGUAAGUGCUGUCCAGUGGAGGCAGCUUAAAGUGCCAGCGCCGCUUGAUGGUCAGGCAGAGCCAGAUGACA ((((((((((...(((...((((((...........))))))..)))(((....).)))))))))))).(((........(((....))).(((((.....)))))...)))........ ( -49.80) >consensus CCACUGGACAUCAGGAGCGGCUGGGCACUGGACUUUCCCAGCAGUCCGCCGUAAGUGCUGUCCAAUGGAGGCAGCUUAAAGUGCCAGCGCCGCUUGAUGGUCAGGCAGAGCCAGAUGACA ...(((..(((((..((((((..(((((((((((........))))).........((((((.......))))))....))))))...)))))))))))..)))................ (-47.08 = -47.12 + 0.04)

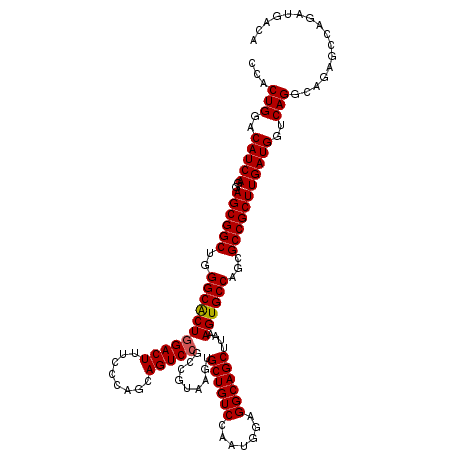
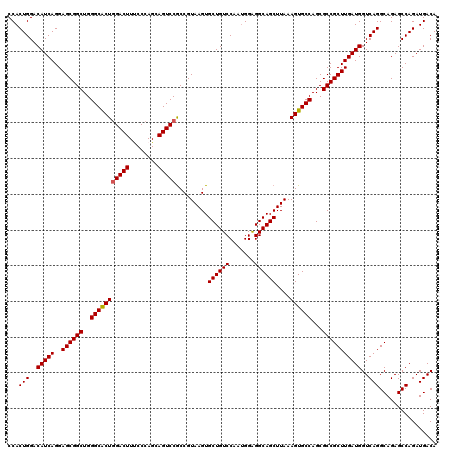
| Location | 13,692,677 – 13,692,797 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -52.88 |
| Consensus MFE | -52.50 |
| Energy contribution | -51.82 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13692677 120 + 27905053 UUUAAGCUGCCUCCAUUGGACAGCACUUACGGUGGACUGCUGGGAAAGUCCAGUGCCCAGCCGCUCCUGAUGUCCAGUGGCCAGCGUACGGGGAGCAACUCGAGCAGCAGUGGCUGCAAC .....((((...((((((((((.((....((((((.(.((((((....))))))).))).)))....)).)))))))))).))))...((((......)))).(((((....)))))... ( -50.50) >DroSec_CAF1 2578 120 + 1 UUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUACUGGGAAAGUCCAGUGCCCAGCCGCUCCUGAUGUCCAGUGGCCAGCGCACGGGGAGCAACUCGAGCAGCAGUGGCUGCAAC .....((((...((((((((((.((....((((((..(((((((....))))))).)).))))....)).)))))))))).))))...((((......)))).(((((....)))))... ( -52.50) >DroSim_CAF1 2593 120 + 1 UUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUGCUGGGAAAGUCCAGUGCCCAGCCGCUCCUGAUGUCCAGUGGCCAGCGCACGGGGAGCAACUCGAGCAGCAGUGGCUGCAAC .....((((...((((((((((.((....((((((.(.((((((....))))))).)).))))....)).)))))))))).))))...((((......)))).(((((....)))))... ( -52.00) >DroEre_CAF1 2599 120 + 1 UUUAAGCUGCCCCCGCUGGACAGCACUUACGGCGGACUGCUGGGCAAGUCCAGCGCCCAGCCGCUCCUGAUGUCCAGUGGCCAGCGCACGGGAAGCAACUCGAGCAGCAGUGGCUGCAAC .....((((...((((((((((.((....((((((..(((((((....))))))).)).))))....)).)))))))))).))))...((((......)))).(((((....)))))... ( -55.90) >DroYak_CAF1 2641 120 + 1 UUUAAGCUGCCUCCACUGGACAGCACUUACGGCGGACUGCUGGGGAAGUCAAGUGCCCAGCCGCUCCUGAUGUCCAGUGGCCAGCGCACGGGGAGCAACUCGAGCAGCAGUGGCUGCAAC .....((((...((((((((((((.......))((((.((((((.(.......).)))))).).)))...)))))))))).))))...((((......)))).(((((....)))))... ( -53.50) >consensus UUUAAGCUGCCUCCAUUGGACAGCACUUACGGCGGACUGCUGGGAAAGUCCAGUGCCCAGCCGCUCCUGAUGUCCAGUGGCCAGCGCACGGGGAGCAACUCGAGCAGCAGUGGCUGCAAC .....((((...((((((((((.((....((((((.(.((((((....))))))).)).))))....)).)))))))))).))))...((((......)))).(((((....)))))... (-52.50 = -51.82 + -0.68)

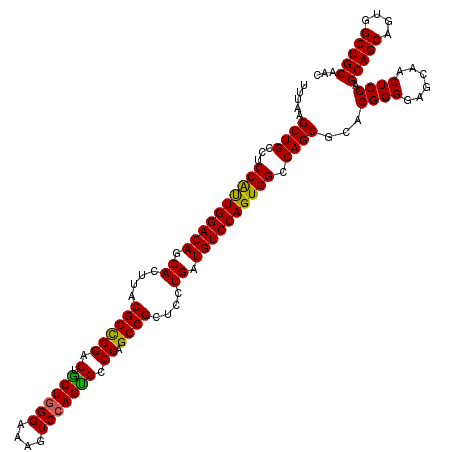
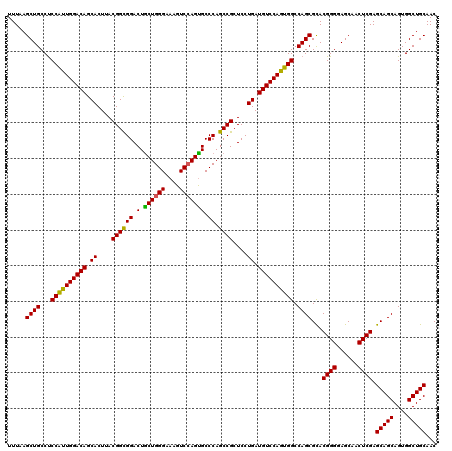
| Location | 13,692,677 – 13,692,797 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -54.16 |
| Consensus MFE | -48.08 |
| Energy contribution | -48.68 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13692677 120 - 27905053 GUUGCAGCCACUGCUGCUCGAGUUGCUCCCCGUACGCUGGCCACUGGACAUCAGGAGCGGCUGGGCACUGGACUUUCCCAGCAGUCCACCGUAAGUGCUGUCCAAUGGAGGCAGCUUAAA ...(((((....)))))..((((((((.((.(....).))(((.((((((.((...((((.(((((.((((......))))..)))))))))...)).)))))).))).))))))))... ( -49.70) >DroSec_CAF1 2578 120 - 1 GUUGCAGCCACUGCUGCUCGAGUUGCUCCCCGUGCGCUGGCCACUGGACAUCAGGAGCGGCUGGGCACUGGACUUUCCCAGUAGUCCGCCGUAAGUGCUGUCCAAUGGAGGCAGCUUAAA ...(((((....)))))..(((((((..((.(((.(....))))((((((.((...(((((.(((((((((......))))).)))))))))...)).))))))..))..)))))))... ( -54.80) >DroSim_CAF1 2593 120 - 1 GUUGCAGCCACUGCUGCUCGAGUUGCUCCCCGUGCGCUGGCCACUGGACAUCAGGAGCGGCUGGGCACUGGACUUUCCCAGCAGUCCGCCGUAAGUGCUGUCCAAUGGAGGCAGCUUAAA ...(((((....)))))..(((((((..((.(((.(....))))((((((.((...(((((.((((.((((......))))..)))))))))...)).))))))..))..)))))))... ( -52.20) >DroEre_CAF1 2599 120 - 1 GUUGCAGCCACUGCUGCUCGAGUUGCUUCCCGUGCGCUGGCCACUGGACAUCAGGAGCGGCUGGGCGCUGGACUUGCCCAGCAGUCCGCCGUAAGUGCUGUCCAGCGGGGGCAGCUUAAA ...(((((....)))))..((((((((.((((.((....))..(((((((.((...(((((.(((((((((......))))).)))))))))...)).)))))))))))))))))))... ( -60.90) >DroYak_CAF1 2641 120 - 1 GUUGCAGCCACUGCUGCUCGAGUUGCUCCCCGUGCGCUGGCCACUGGACAUCAGGAGCGGCUGGGCACUUGACUUCCCCAGCAGUCCGCCGUAAGUGCUGUCCAGUGGAGGCAGCUUAAA ...(((((....)))))..((((((((.((.(....).))((((((((((...(((...((((((...........))))))..)))(((....).)))))))))))).))))))))... ( -53.20) >consensus GUUGCAGCCACUGCUGCUCGAGUUGCUCCCCGUGCGCUGGCCACUGGACAUCAGGAGCGGCUGGGCACUGGACUUUCCCAGCAGUCCGCCGUAAGUGCUGUCCAAUGGAGGCAGCUUAAA ...(((((....)))))..(((((((..((.(((.(....))))((((((.((...(((((.((((.((((......))))..)))))))))...)).))))))..))..)))))))... (-48.08 = -48.68 + 0.60)

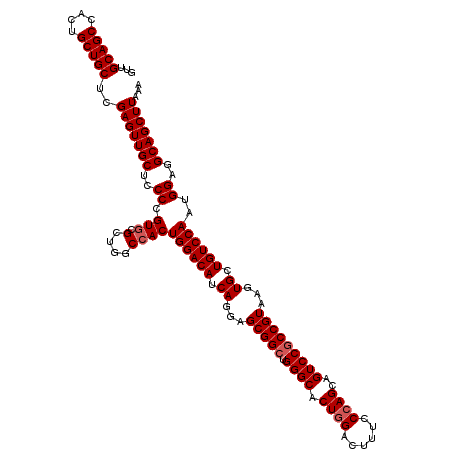
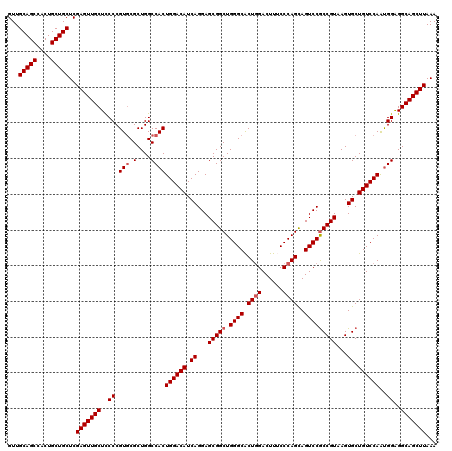
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:55 2006