
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,682,361 – 13,682,608 |
| Length | 247 |
| Max. P | 0.999913 |

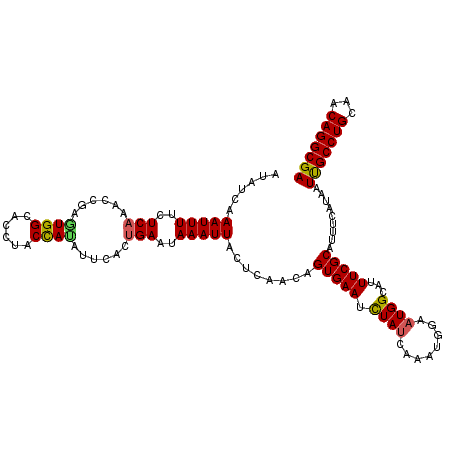
| Location | 13,682,361 – 13,682,467 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13682361 106 + 27905053 AGUUUUUUUAUUUUUAUUAGUGAAUAGAUUUUUUAAUAAUGGCC-CGGGCCCCAGGGAUGUGUACACCAAAUUGUCGGACAUGCAGUUGGACUGGUGUGCGGUCAAC (((((.((((((......)))))).))))).........(((((-....((....))....((((((((....(((.(((.....))).)))))))))))))))).. ( -29.00) >DroSec_CAF1 9689 100 + 1 AGUUU-------UUUAUUUGCGGAUAGAUUUUUUAAUAAUGGCCCCGGGCCCCAGGGACGUGUACACCAAAUUGUCCGACAUGCAGUUGGACUGGUGUGCGGUCAAC .....-------((((((....))))))...........(((((.((..((....)).)).((((((((....(((((((.....)))))))))))))))))))).. ( -32.60) >DroSim_CAF1 9740 102 + 1 AGUUUUU-----UUUAUUUGCGAAUAGAUUUUUUAAUAAUGGCCCCGGGCCCCAGGGACGUGUACACCAAAUUGUCCGACAUGCAGUUGGACUGGUGUGCGGUCAAC .......-----((((((....))))))...........(((((.((..((....)).)).((((((((....(((((((.....)))))))))))))))))))).. ( -32.10) >DroEre_CAF1 4168 99 + 1 CCUU--------UUUAUUUGUGGAUAGAUUUUUUAAUAAUGGCCCCGCGGCCCAGGGACGUGUACACCAAAUUGUUCGACAUGCAGUUGGACUGGUGUGCGGUCAAC (((.--------((((((....))))))............((((....)))).)))(((.(((((((((....(((((((.....)))))))))))))))))))... ( -32.10) >DroYak_CAF1 4367 99 + 1 CGUU--------UUUAUUUGUGGAUAGAUUUUUUAAUAAUGGCCCCAGGCGCAAAGGACGUGUACACCAAAUUGUCCGACAUGCAGUUGAACUGGUGUGCGGUCAAC ....--------((((((....))))))...........(((((...))).))...(((.(((((((((....((.((((.....)))).))))))))))))))... ( -23.30) >consensus AGUUU_______UUUAUUUGUGGAUAGAUUUUUUAAUAAUGGCCCCGGGCCCCAGGGACGUGUACACCAAAUUGUCCGACAUGCAGUUGGACUGGUGUGCGGUCAAC ............((((((....))))))............(((.....))).....(((.(((((((((....(((((((.....)))))))))))))))))))... (-25.28 = -25.56 + 0.28)

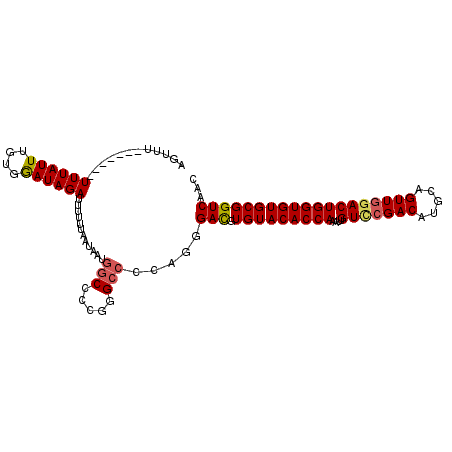

| Location | 13,682,400 – 13,682,492 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -36.56 |
| Energy contribution | -36.72 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13682400 92 + 27905053 UGGCC-CGGGCCCCAGGGAUGUGUACACCAAAUUGUCGGACAUGCAGUUGGACUGGUGUGCGGUCAACCAAGGAUGUCGUCCCUUUUGGCCAG .....-..((((..(((((((.((((((((....(((.(((.....))).)))))))))))((....))........)))))))...)))).. ( -38.30) >DroSec_CAF1 9721 93 + 1 UGGCCCCGGGCCCCAGGGACGUGUACACCAAAUUGUCCGACAUGCAGUUGGACUGGUGUGCGGUCAACCAUGGAUAUCGUCCCUUUUGGCCAG ........((((..(((((((.((((((((....(((((((.....)))))))))))))))((....))........)))))))...)))).. ( -43.80) >DroSim_CAF1 9774 93 + 1 UGGCCCCGGGCCCCAGGGACGUGUACACCAAAUUGUCCGACAUGCAGUUGGACUGGUGUGCGGUCAACCAUGGAUAUCGUCCCUUUUGGCCAG ........((((..(((((((.((((((((....(((((((.....)))))))))))))))((....))........)))))))...)))).. ( -43.80) >DroEre_CAF1 4199 93 + 1 UGGCCCCGCGGCCCAGGGACGUGUACACCAAAUUGUUCGACAUGCAGUUGGACUGGUGUGCGGUCAACCAAGGAUGUCGUCCCUUUUGGCCAG .........((((.(((((((.((((((((....(((((((.....)))))))))))))))((....))........)))))))...)))).. ( -40.80) >DroYak_CAF1 4398 93 + 1 UGGCCCCAGGCGCAAAGGACGUGUACACCAAAUUGUCCGACAUGCAGUUGAACUGGUGUGCGGUCAACCAAGGAUAUUGUCCCUUUUGGCCAG (((((..(((.((((..(((.(((((((((....((.((((.....)))).))))))))))))))..(....)...)))).)))...))))). ( -33.30) >consensus UGGCCCCGGGCCCCAGGGACGUGUACACCAAAUUGUCCGACAUGCAGUUGGACUGGUGUGCGGUCAACCAAGGAUAUCGUCCCUUUUGGCCAG (((((.........(((((((.((((((((....(((((((.....)))))))))))))))((....))........)))))))...))))). (-36.56 = -36.72 + 0.16)



| Location | 13,682,400 – 13,682,492 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -29.96 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13682400 92 - 27905053 CUGGCCAAAAGGGACGACAUCCUUGGUUGACCGCACACCAGUCCAACUGCAUGUCCGACAAUUUGGUGUACACAUCCCUGGGGCCCG-GGCCA ((((((...(((((((((.......))))...(.(((((((((..((.....))..)))....)))))).)...)))))..)).)))-).... ( -29.10) >DroSec_CAF1 9721 93 - 1 CUGGCCAAAAGGGACGAUAUCCAUGGUUGACCGCACACCAGUCCAACUGCAUGUCGGACAAUUUGGUGUACACGUCCCUGGGGCCCGGGGCCA ((((((...(((((((........((....))(.((((((((((.((.....)).))))....)))))).).)))))))..)).))))..... ( -38.30) >DroSim_CAF1 9774 93 - 1 CUGGCCAAAAGGGACGAUAUCCAUGGUUGACCGCACACCAGUCCAACUGCAUGUCGGACAAUUUGGUGUACACGUCCCUGGGGCCCGGGGCCA ((((((...(((((((........((....))(.((((((((((.((.....)).))))....)))))).).)))))))..)).))))..... ( -38.30) >DroEre_CAF1 4199 93 - 1 CUGGCCAAAAGGGACGACAUCCUUGGUUGACCGCACACCAGUCCAACUGCAUGUCGAACAAUUUGGUGUACACGUCCCUGGGCCGCGGGGCCA ..((((...((((((((((((...(((((..((.(((.(((.....)))..)))))..))))).)))))...))))))).))))......... ( -33.50) >DroYak_CAF1 4398 93 - 1 CUGGCCAAAAGGGACAAUAUCCUUGGUUGACCGCACACCAGUUCAACUGCAUGUCGGACAAUUUGGUGUACACGUCCUUUGCGCCUGGGGCCA ..((((..(((((((.........((....))(.((((((((((.((.....)).))))....)))))).)..)))))))........)))). ( -29.10) >consensus CUGGCCAAAAGGGACGAUAUCCUUGGUUGACCGCACACCAGUCCAACUGCAUGUCGGACAAUUUGGUGUACACGUCCCUGGGGCCCGGGGCCA ..((((...(((((((........((....))(.((((((((((.((.....)).))))....)))))).).))))))).(....)..)))). (-29.96 = -30.32 + 0.36)

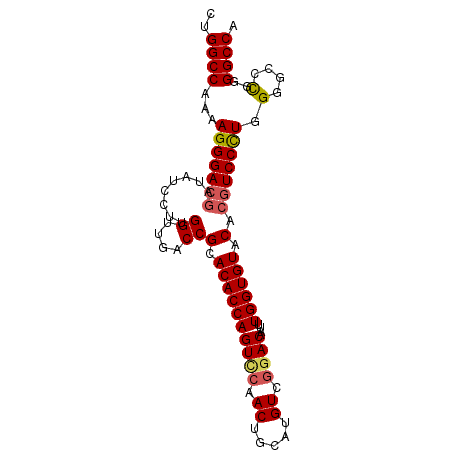

| Location | 13,682,492 – 13,682,608 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.50 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13682492 116 - 27905053 AUAUCAAAUUUUCUCAAAGCGAGUGGCACCUACCAUAUUCACCGAAUAAAUUACUCAACAGUGAAUUUAUCAAAUGGAAUGGCAUUUCGCAUUUCAUAAUUGCCUGCAACAGGCGA ..................(((((((.((....((((((((...)))))..(((((....)))))..........)))..)).)).))))).........(((((((...))))))) ( -22.10) >DroSec_CAF1 9814 116 - 1 AUAUCAAAUUUUCUCAAACCGAAUGGCACCUACCAUAUUCACUGAAUAAAUUACUCGACAGUGAAUCUAUCAAAUGGUAUGGCAUUUCGCAUUUCAUAAUUGCCUGCAACAGGCGA ...................((((..((.(.((((((((((((((..............)))))))).......)))))).)))..))))..........(((((((...))))))) ( -25.65) >DroSim_CAF1 9867 116 - 1 AUAUCAAAUUUUCUCAAACCGAGUGGCACCUACCAUAUUCACUGAAUAAAUUACUCAAUAGUGAAUCUAUCAACUGGUUUGGCAUUUCGCAUUUCAUAAUCGCCUGCAACAGGCGA ..............(((((((.((((......))))((((((((..............))))))))........)))))))((.....)).........(((((((...))))))) ( -26.34) >DroEre_CAF1 4292 114 - 1 AUAUCAAAUUUUCUCAAACCGAGUGGCAUCCACUA--CUCACUGAAUAAAUUACCCAACUGUGAAUCUAUCAAACGGAAUGGCAUUUCGCAUUUCAUAAUUGCCUGCAACAGGCGA ......(((((..(((....((((((......)))--)))..)))..))))).......((((((.((((........))))...))))))........(((((((...))))))) ( -22.10) >DroYak_CAF1 4491 116 - 1 AUAUCAAAUUUUCUCAAACCAAGUGGCAUCAACUACACUCACUGAAUAAAUUACUCGACUGUGAAUCUAUAAAAUGGGAUGGCAUUUCGCAUUUCAUAAUUGCCUGCAACAGGCGA ......(((((..(((.....((((..........))))...)))..)))))....((.((((((.((((........))))...))))))..))....(((((((...))))))) ( -18.80) >consensus AUAUCAAAUUUUCUCAAACCGAGUGGCACCUACCAUAUUCACUGAAUAAAUUACUCAACAGUGAAUCUAUCAAAUGGAAUGGCAUUUCGCAUUUCAUAAUUGCCUGCAACAGGCGA ......(((((..(((......((((......))))......)))..)))))........(((((.((((........))))...))))).........(((((((...))))))) (-16.70 = -16.50 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:42 2006