
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,679,968 – 13,680,164 |
| Length | 196 |
| Max. P | 0.989635 |

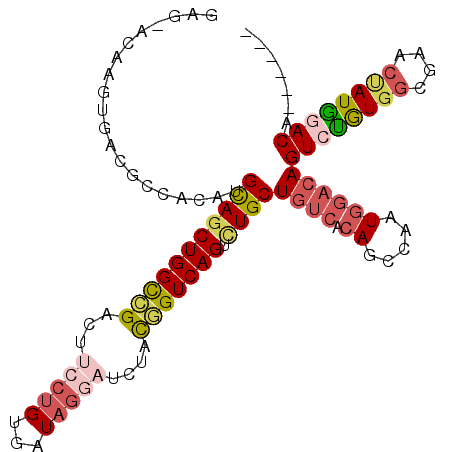
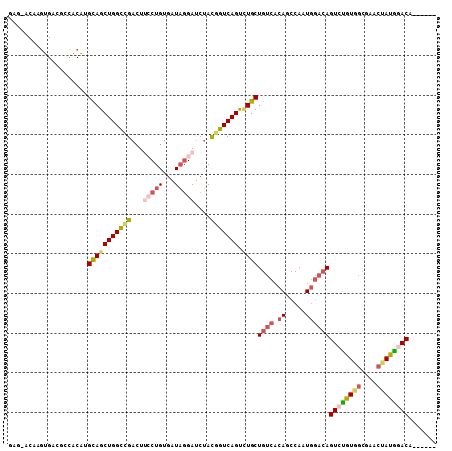
| Location | 13,679,968 – 13,680,070 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.58 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.95 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13679968 102 - 27905053 GAGAGCAAGUGACGCCACAUGCAGCUGGCCGACUUCCUGUGAUAGGAUCUACGGUCAGUCUGCUGUCACAACCAAUGGACAGUCUGUGGCGAACUAUGGACA------ .....(((((..(((((((.(((((((((((...(((((...)))))....))))))).))))((((.((.....))))))...))))))).))).))....------ ( -39.90) >DroSec_CAF1 7287 102 - 1 GAGGACAAGUGACGCCACAUGCAGCUGGCCGACUUCCUGUGGUAGGAUCUACGGUCAGUCUGCUGUCACAGCCAAUGGACAGUCUGUGGCGAACUAUGGACA------ ..(..(((((..(((((((.(((((((((((...(((((...)))))....))))))).))))((((.((.....))))))...))))))).))).))..).------ ( -43.70) >DroSim_CAF1 7320 102 - 1 GAGGACAAGUGACGCCACAUGCAGCUGGCCGACUUCCUGUGAUAGGAUCUACGGUCAGUCUGCUGUCACAGCCAAUGGACAGUCUGUGGCGAACUAUGGACA------ ..(..(((((..(((((((.(((((((((((...(((((...)))))....))))))).))))((((.((.....))))))...))))))).))).))..).------ ( -43.70) >DroEre_CAF1 1838 81 - 1 -------------------UGCAGCUGGCCGACU--CUGUGAUAGGAUCUACGGUCAGUGUGCUGUCACAGCCAAUGGACAGUCUGUGGCGGACUAUAGACU------ -------------------.(((((((((((..(--((......)))....)))))))).)))((((.((.....))))))((((((((....)))))))).------ ( -29.40) >DroYak_CAF1 1834 81 - 1 -------------------UGCAGCUGGCAGAGU--CUGUGAUAGGAUCUACGGUCAGUCUGCUGUCGCAACCAAUGGACAGUCUGUGGCGGACCAUAUACC------ -------------------.(((((((((..(((--((......))).))...))))).))))((((.((.....))))))((.(((((....))))).)).------ ( -24.40) >DroAna_CAF1 6949 102 - 1 AACAACAAAGGACUCUCCCUGUAGCUGGUUGCCUUCCUGCGAUUCGAUCUAUAGUCAGCUUGCUGUCACAUCCAAUCCUGAGUCCAU------AUAUGUACGGAAGUA .........((((((...(((((((..(((((......)))))..)..)))))).(((....)))..............))))))..------.....(((....))) ( -24.00) >consensus GAG_ACAAGUGACGCCACAUGCAGCUGGCCGACUUCCUGUGAUAGGAUCUACGGUCAGUCUGCUGUCACAGCCAAUGGACAGUCUGUGGCGAACUAUGGACA______ ....................(((((((((((...(((((...)))))....))))))).))))((((.((.....))))))((((((((....))))))))....... (-22.64 = -23.95 + 1.31)

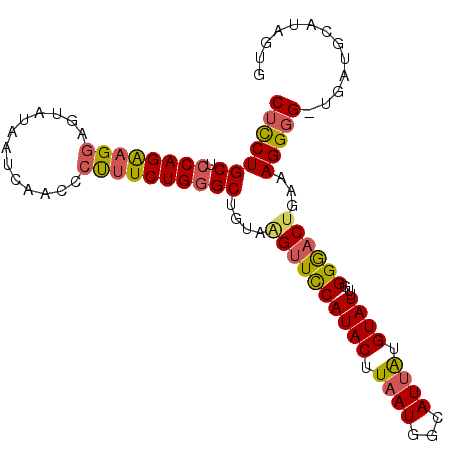
| Location | 13,680,070 – 13,680,164 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.36 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13680070 94 + 27905053 CACUAUGCAUCAUCCCCUUUCAGUCCCCAAAUACAUAAUGCCAUUAAGUAUGAAACUUACAGCCCAGAAGGGUUGGA------UCCUUCUGGAGCAGGAG ............(((.............................(((((.....)))))..((((((((((((...)------))))))))).)).))). ( -22.00) >DroSec_CAF1 7389 99 + 1 CACUAUGCAUCA-CCCCUUUCAGAUCCCAAAUACAUAAUGCCAUUAAGUAUGGAGCCUACAGCCCAGAAAGGGUUGAUUAUACUCCUUCUGGAGCAGGAG ...((((.((((-.(((((((.........((((.((((...)))).))))((.((.....)))).))))))).))))))))(((((.(....).))))) ( -25.50) >DroSim_CAF1 7422 99 + 1 CACUAUGCAUCA-CCCCUUUUAGUUCCCAAAUACAUAAUGCCAUUAAGUAUGGAACUUACAGCCCAGAAAGGGUUGACUAUACUCCUUCUGGAGCAGGAG ............-..(((...((((((...((((.((((...)))).))))))))))....((((((((.((((.......)))).)))))).))))).. ( -24.90) >DroEre_CAF1 1919 89 + 1 ----------CA-CCCCUUUCAGUCCCCAAAUACACAAUUCCAUUAAGUAUGGAACUUACUGCCCAGAAAGGGGGGAUUAUACUCCUCCUGGAGCAGAAG ----------..-.........................((((((.....))))))(((.((((((((...((((((......)))))))))).))))))) ( -25.00) >DroYak_CAF1 1915 88 + 1 ----------CA-CCCCUUUCGGUCCCCAAAUACACGAUUUCAUUAAGUAUGGAACUGAGAGCCCAGAAAGGGGUGAUUAUACUUC-CCUGUAGCAGAUG ----------((-((((((((((...)).......((.((((((.....)))))).))........))))))))))....(((...-...)))....... ( -22.60) >consensus CACUAUGCAUCA_CCCCUUUCAGUCCCCAAAUACAUAAUGCCAUUAAGUAUGGAACUUACAGCCCAGAAAGGGUUGAUUAUACUCCUUCUGGAGCAGGAG ..............(((((((.((........))......((((.....)))).............))))))).........((((....))))...... (-11.80 = -12.36 + 0.56)

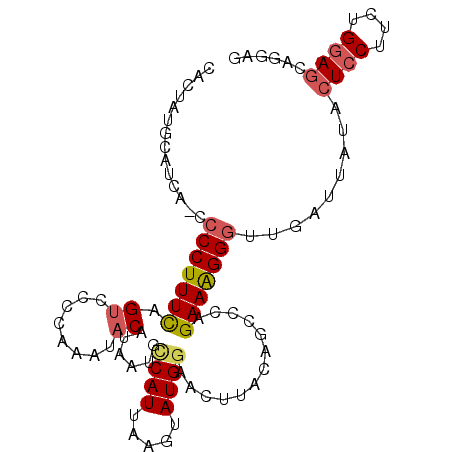

| Location | 13,680,070 – 13,680,164 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13680070 94 - 27905053 CUCCUGCUCCAGAAGGA------UCCAACCCUUCUGGGCUGUAAGUUUCAUACUUAAUGGCAUUAUGUAUUUGGGGACUGAAAGGGGAUGAUGCAUAGUG ..((.((.((((((((.------......))))))))))..(((((.....)))))..))(((((((((((......((....))....))))))))))) ( -30.10) >DroSec_CAF1 7389 99 - 1 CUCCUGCUCCAGAAGGAGUAUAAUCAACCCUUUCUGGGCUGUAGGCUCCAUACUUAAUGGCAUUAUGUAUUUGGGAUCUGAAAGGGG-UGAUGCAUAGUG ..((((((((((((((.((.......)).)))))))))..)))))..((((.....))))(((((((((((.....(((....))).-.))))))))))) ( -31.70) >DroSim_CAF1 7422 99 - 1 CUCCUGCUCCAGAAGGAGUAUAGUCAACCCUUUCUGGGCUGUAAGUUCCAUACUUAAUGGCAUUAUGUAUUUGGGAACUAAAAGGGG-UGAUGCAUAGUG (((((((.((((((((.((.......)).))))))))))....((((((((((.((((...)))).))))...))))))...)))))-............ ( -27.90) >DroEre_CAF1 1919 89 - 1 CUUCUGCUCCAGGAGGAGUAUAAUCCCCCCUUUCUGGGCAGUAAGUUCCAUACUUAAUGGAAUUGUGUAUUUGGGGACUGAAAGGGG-UG---------- ....((((((....))))))......((((((((...(((...((((((((.....)))))))).)))....(....).))))))))-..---------- ( -31.00) >DroYak_CAF1 1915 88 - 1 CAUCUGCUACAGG-GAAGUAUAAUCACCCCUUUCUGGGCUCUCAGUUCCAUACUUAAUGAAAUCGUGUAUUUGGGGACCGAAAGGGG-UG---------- ..((((...))))-..........((((((((((.((..((((((....((((...........))))..)))))).))))))))))-))---------- ( -25.20) >consensus CUCCUGCUCCAGAAGGAGUAUAAUCAACCCUUUCUGGGCUGUAAGUUCCAUACUUAAUGGCAUUAUGUAUUUGGGGACUGAAAGGGG_UGAUGCAUAGUG (((((((.((((((((.............))))))))))....((((((((((.((((...)))).))))...))))))...)))))............. (-19.52 = -19.52 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:37 2006