
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,673,641 – 13,673,751 |
| Length | 110 |
| Max. P | 0.764249 |

| Location | 13,673,641 – 13,673,751 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -17.66 |
| Energy contribution | -18.89 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13673641 110 + 27905053 GAUGGGGCAGGGGUGUGGAACACGUAUUAACUCGACACAUGUGUAAAUCCAAUUCAAAACAUGCCACGUGCAACCA--------CCCUGCCCCGCAUUUUCCACUCCCCCCCCCCCCC ...((((..((((.(((((((((((.............))))))................((((...(.(((....--------...))))..))))..))))).))))..))))... ( -32.92) >DroEre_CAF1 42438 110 + 1 GAGGGGGCAGAGGGGUGUAACACGUAUUAACUCGACACAUGUGUAAAUCCAAUUCAAAACAUGCCACGUGCAACCAAGCUACCACCCUGCCCCCCGUUUUCCA-------CA-CCCAC (.((((((((.(((((((.((((((.............)))))).................(((.....))).....)).))).)))))))))))........-------..-..... ( -31.12) >DroYak_CAF1 39853 111 + 1 GAUGGUGCAGAGGGGUGCAACACGUAUUAACUCGACACAUGUGUAAAUCCAAUUCAAAACAUGCCACGUGCAACCAACUUACCACCCCGACCUCCGUUCUCCA-------UCCUCCAC (((((.((.((((((((..((((((.............)))))).................(((.....)))..........))))))....)).))...)))-------))...... ( -23.02) >consensus GAUGGGGCAGAGGGGUGCAACACGUAUUAACUCGACACAUGUGUAAAUCCAAUUCAAAACAUGCCACGUGCAACCAA__UACCACCCUGCCCCCCGUUUUCCA_______CCCCCCAC ..((((((((..((((...((((((.............))))))..))))...........(((.....)))..............))))))))........................ (-17.66 = -18.89 + 1.23)

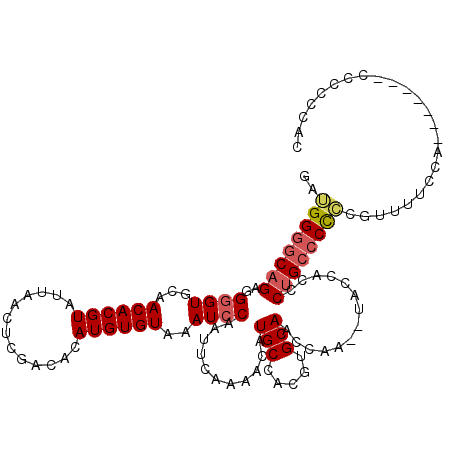

| Location | 13,673,641 – 13,673,751 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -27.55 |
| Energy contribution | -29.33 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13673641 110 - 27905053 GGGGGGGGGGGGGAGUGGAAAAUGCGGGGCAGGG--------UGGUUGCACGUGGCAUGUUUUGAAUUGGAUUUACACAUGUGUCGAGUUAAUACGUGUUCCACACCCCUGCCCCAUC ...((((.(((((.(((((....(((..((((..--------...)))).))).((((((....((((.(((..((....))))).))))...))))))))))).))))).))))... ( -45.40) >DroEre_CAF1 42438 110 - 1 GUGGG-UG-------UGGAAAACGGGGGGCAGGGUGGUAGCUUGGUUGCACGUGGCAUGUUUUGAAUUGGAUUUACACAUGUGUCGAGUUAAUACGUGUUACACCCCUCUGCCCCCUC .....-..-------........(((((((((((.(((.((......))..(((((((((....((((.(((..((....))))).))))...)))))))))))).))))))))))). ( -44.50) >DroYak_CAF1 39853 111 - 1 GUGGAGGA-------UGGAGAACGGAGGUCGGGGUGGUAAGUUGGUUGCACGUGGCAUGUUUUGAAUUGGAUUUACACAUGUGUCGAGUUAAUACGUGUUGCACCCCUCUGCACCAUC ......((-------(((....((((....(((((.((((.....))))..(..((((((....((((.(((..((....))))).))))...))))))..))))))))))..))))) ( -31.90) >consensus GUGGGGGG_______UGGAAAACGGGGGGCAGGGUGGUA__UUGGUUGCACGUGGCAUGUUUUGAAUUGGAUUUACACAUGUGUCGAGUUAAUACGUGUUACACCCCUCUGCCCCAUC .........................(((((((((.(((.....(......)(((((((((....((((.(((..((....))))).))))...)))))))))))).)))))))))... (-27.55 = -29.33 + 1.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:30 2006