| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,671,368 – 13,671,497 |
| Length | 129 |
| Max. P | 0.951952 |

| Location | 13,671,368 – 13,671,458 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -13.52 |
| Energy contribution | -15.15 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13671368 90 + 27905053 AAUCCUGGCCAAGUCACAUCAUUCUGUUUUCCAUAUACUCGUAUGUACAUAUACGAAUAUACGAGUAUA------------------UAUGAUUAUUUGUAUAGGCUU ...((((..(((((...(((((..........((((((((((((((.(......).)))))))))))))------------------)))))).)))))..))))... ( -23.80) >DroSim_CAF1 36533 108 + 1 AAUCCCGGCCAAGUCACAUCACUCUGUUUUCCAUAUACUCGUAUGUGCAUAAACGAAUAUACGAGUAUACACAUACAUAUAUUUGUCUAUGAUUGUUUGUAUAGGCUU ......((((.......................(((((((((((((.(......).)))))))))))))...(((((.(((.((......)).))).))))).)))). ( -24.60) >DroEre_CAF1 40229 100 + 1 AAUCCUGGCCAAGUCACAUCAUGCUGUUUUCAAUACUCGUAUAUGUACAUAUACGAAUAAACGAGUAUACUCGU--------AUGUCUAUGACUGUUUGUAUAGGCCU ......((((..((.((((.((((.((.......))..)))))))))).((((((((((.(((((....)))))--------..(((...))))))))))))))))). ( -27.30) >DroYak_CAF1 37540 94 + 1 AAUCCUGGCCAAGUCACAUCAUGCUGUUUUCCAUAUACGAGUAUGUACAUAUAGGAAUAUACGAGUAU--------------AUGUCUAUGAUUGUUUGUAUAGGCCU ...((((..((((.((.((((((........(((((((..((((((.(......).))))))..))))--------------)))..))))))))))))..))))... ( -20.90) >consensus AAUCCUGGCCAAGUCACAUCAUGCUGUUUUCCAUAUACGCGUAUGUACAUAUACGAAUAUACGAGUAUA_____________AUGUCUAUGAUUGUUUGUAUAGGCCU ......((((.......................(((((((((((((.(......).)))))))))))))..................((..(....)..))..)))). (-13.52 = -15.15 + 1.63)


| Location | 13,671,368 – 13,671,458 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -12.99 |
| Energy contribution | -15.05 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13671368 90 - 27905053 AAGCCUAUACAAAUAAUCAUA------------------UAUACUCGUAUAUUCGUAUAUGUACAUACGAGUAUAUGGAAAACAGAAUGAUGUGACUUGGCCAGGAUU ..(((....((....(((((.------------------.....((((((((((((((......))))))))))))))........))))).))....)))....... ( -21.44) >DroSim_CAF1 36533 108 - 1 AAGCCUAUACAAACAAUCAUAGACAAAUAUAUGUAUGUGUAUACUCGUAUAUUCGUUUAUGCACAUACGAGUAUAUGGAAAACAGAGUGAUGUGACUUGGCCGGGAUU ..(((....((....(((((......((((.((((((((((((..((......))..)))))))))))).)))).((.....))..))))).))....)))....... ( -26.70) >DroEre_CAF1 40229 100 - 1 AGGCCUAUACAAACAGUCAUAGACAU--------ACGAGUAUACUCGUUUAUUCGUAUAUGUACAUAUACGAGUAUUGAAAACAGCAUGAUGUGACUUGGCCAGGAUU .((((.........(((((((..(((--------(((((....))))).(((((((((((....)))))))))))...........))).))))))).))))...... ( -31.10) >DroYak_CAF1 37540 94 - 1 AGGCCUAUACAAACAAUCAUAGACAU--------------AUACUCGUAUAUUCCUAUAUGUACAUACUCGUAUAUGGAAAACAGCAUGAUGUGACUUGGCCAGGAUU .((((....((....(((((.....(--------------(((....))))((((((((((........)))))).))))......))))).))....))))...... ( -20.40) >consensus AAGCCUAUACAAACAAUCAUAGACAU_____________UAUACUCGUAUAUUCGUAUAUGUACAUACGAGUAUAUGGAAAACAGAAUGAUGUGACUUGGCCAGGAUU .((((....((....(((((.........................(((((((((((((......)))))))))))))(....)...))))).))....))))...... (-12.99 = -15.05 + 2.06)

| Location | 13,671,396 – 13,671,497 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -15.45 |
| Energy contribution | -17.32 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13671396 101 + 27905053 UUCCAUAUACUCGUAUGUACAUAUACGAAUAUACGAGUAUA------------------UAUGAUUAUUUGUAUAGGCUUAACGUGCCGAAAAUGUGUUUAUGAAUCAAUAUUGCGCAU (((.((((((((((((((.(......).)))))))))))))------------------)...............(((.......)))))).((((((.(((......)))..)))))) ( -26.30) >DroSim_CAF1 36561 118 + 1 UUCCAUAUACUCGUAUGUGCAUAAACGAAUAUACGAGUAUACACAUACAUAUAUUUGUCUAUGAUUGUUUGUAUAGGCUUAACGUGCCGA-AAUGUGUUUAUGAAUCAAUAUUGCGCAU .....(((((((((((((.(......).)))))))))))))...(((((.(((.((......)).))).))))).(((.......)))..-.((((((.(((......)))..)))))) ( -28.40) >DroEre_CAF1 40257 110 + 1 UUCAAUACUCGUAUAUGUACAUAUACGAAUAAACGAGUAUACUCGU--------AUGUCUAUGACUGUUUGUAUAGGCCUAACGUGCCGA-AAUGUGUUUAUGAAUCAAUAUUGCGCAC (((.((((((((((((....))))))))(((.(((((....)))))--------.)))............)))).((((....).)))))-).(((((.(((......)))..))))). ( -26.90) >DroYak_CAF1 37568 104 + 1 UUCCAUAUACGAGUAUGUACAUAUAGGAAUAUACGAGUAU--------------AUGUCUAUGAUUGUUUGUAUAGGCCUAACGUGCCGA-AAUGUGUUUAUGAAUCAAUAUUGCGCAC ((((((((((..((((((.(......).))))))..))))--------------)))..................((((....).)))))-).(((((.(((......)))..))))). ( -23.10) >consensus UUCCAUAUACGCGUAUGUACAUAUACGAAUAUACGAGUAUA_____________AUGUCUAUGAUUGUUUGUAUAGGCCUAACGUGCCGA_AAUGUGUUUAUGAAUCAAUAUUGCGCAC .....(((((((((((((.(......).)))))))))))))..................................(((.......))).....(((((.(((......)))..))))). (-15.45 = -17.32 + 1.88)

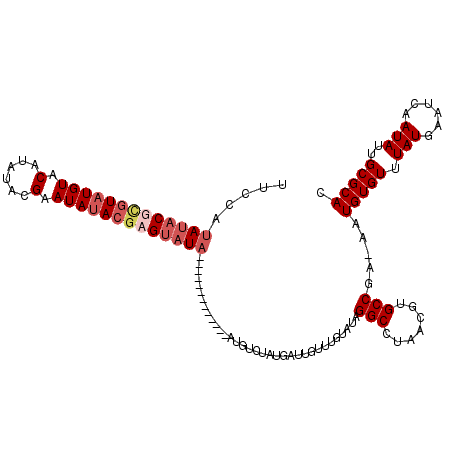
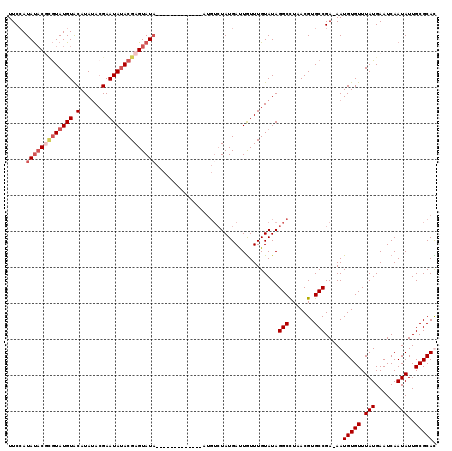
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:28 2006