| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
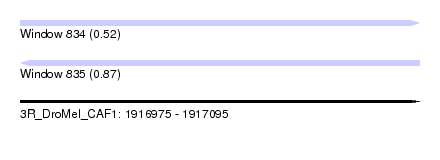
| Location | 1,916,975 – 1,917,095 |
| Length | 120 |
| Max. P | 0.868621 |

| Location | 1,916,975 – 1,917,095 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -19.31 |
| Energy contribution | -20.50 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
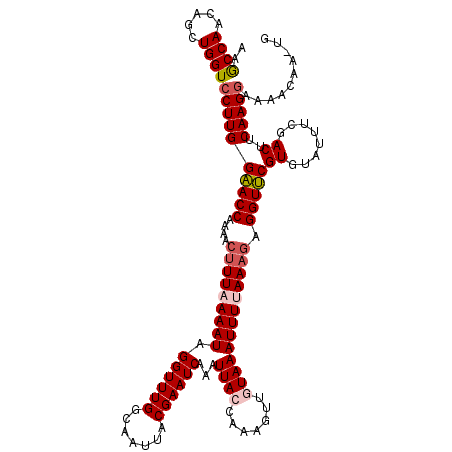
>3R_DroMel_CAF1 1916975 120 + 27905053 AAACCAACAGCUGGUCCUUGGAACCAAAACUUUAAAAUGGGUUUGGCAAUUACGAAUCAAAUUACAAAAGUUGUAAAUUUAAAAUAGGUUCGUGUAUUUCGACUUCAAGGUAAACAAAUG ..((((.....))))(((((((.(((((.(((......))))))))....((((((((((((((((.....))).)))))......)))))))).........))))))).......... ( -22.10) >DroSec_CAF1 75375 120 + 1 AAGCCAACAGCUGGUCCUUGGAACCAAAACUUUAAAAUAGGUUUGGCAAUUACGAAUCAAAUUACCAAAGUUUUAAAUUUUAAAGAGGUUCGUGUAUUUUGACUUCAAGGAAAACAACUG .......(((.((.(((((((((((....(((((((((((.(((((.((((........)))).))))).))....))))))))).)))))((........))..))))))...)).))) ( -27.40) >DroSim_CAF1 76151 120 + 1 AAGCCAACAGCUGGUCCUUGGAACCAAAACUUUAAAAUAGGUUUGGCAAUUACGAAUCAAAUUACCAAAGUUUUAAAUUUUAAAGAGGUUCGUGUAUUUUGACUUCAAGGAAAACAAGUG .........((((.(((((((((((....(((((((((((.(((((.((((........)))).))))).))....))))))))).)))))((........))..))))))...).))). ( -25.10) >DroEre_CAF1 77329 109 + 1 AAGCCAACAUCUGGUGCUUGGCACCGAAACUUUAAAAUAGGUUUGGCAAUUACGAAUCAAAUUACCAAAGUUGUAAAUUUGAAUGAGGUGCGUAUAUUUCGACUACAAG----------- ..((((.....)))).(((((((((.....((((((((((.(((((.((((........)))).))))).))))...))))))...)))))((........))..))))----------- ( -22.40) >consensus AAGCCAACAGCUGGUCCUUGGAACCAAAACUUUAAAAUAGGUUUGGCAAUUACGAAUCAAAUUACCAAAGUUGUAAAUUUUAAAGAGGUUCGUGUAUUUCGACUUCAAGGAAAACAA_UG ..((((.....))))((((((((((....(((((((((.((((((.......))))))...((((.......))))))))))))).)))))((........))..))))).......... (-19.31 = -20.50 + 1.19)

| Location | 1,916,975 – 1,917,095 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -20.17 |
| Energy contribution | -21.30 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
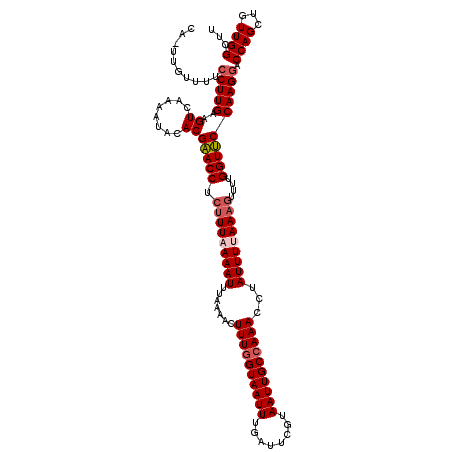
>3R_DroMel_CAF1 1916975 120 - 27905053 CAUUUGUUUACCUUGAAGUCGAAAUACACGAACCUAUUUUAAAUUUACAACUUUUGUAAUUUGAUUCGUAAUUGCCAAACCCAUUUUAAAGUUUUGGUUCCAAGGACCAGCUGUUGGUUU ..........(((((..((......))..(((((.((((((((........(((.((((((........)))))).))).....))))))))...))))))))))(((((...))))).. ( -19.62) >DroSec_CAF1 75375 120 - 1 CAGUUGUUUUCCUUGAAGUCAAAAUACACGAACCUCUUUAAAAUUUAAAACUUUGGUAAUUUGAUUCGUAAUUGCCAAACCUAUUUUAAAGUUUUGGUUCCAAGGACCAGCUGUUGGCUU ((((((...((((((..((......))..(((((.(((((((((.......((((((((((........))))))))))...)))))))))....))))))))))).))))))....... ( -33.50) >DroSim_CAF1 76151 120 - 1 CACUUGUUUUCCUUGAAGUCAAAAUACACGAACCUCUUUAAAAUUUAAAACUUUGGUAAUUUGAUUCGUAAUUGCCAAACCUAUUUUAAAGUUUUGGUUCCAAGGACCAGCUGUUGGCUU .........((((((..((......))..(((((.(((((((((.......((((((((((........))))))))))...)))))))))....)))))))))))..(((.....))). ( -28.70) >DroEre_CAF1 77329 109 - 1 -----------CUUGUAGUCGAAAUAUACGCACCUCAUUCAAAUUUACAACUUUGGUAAUUUGAUUCGUAAUUGCCAAACCUAUUUUAAAGUUUCGGUGCCAAGCACCAGAUGUUGGCUU -----------..((..((((.......)).))..))..........((((((((((((((........))))))))))...........((((.(((((...))))))))))))).... ( -20.90) >consensus CA_UUGUUUUCCUUGAAGUCAAAAUACACGAACCUCUUUAAAAUUUAAAACUUUGGUAAUUUGAUUCGUAAUUGCCAAACCUAUUUUAAAGUUUUGGUUCCAAGGACCAGCUGUUGGCUU ..........(((((..((........))(((((.(((((((((.......((((((((((........))))))))))...)))))))))....)))))))))).((((...))))... (-20.17 = -21.30 + 1.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:10 2006