
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,662,903 – 13,662,994 |
| Length | 91 |
| Max. P | 0.994841 |

| Location | 13,662,903 – 13,662,994 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -12.73 |
| Energy contribution | -13.36 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13662903 91 + 27905053 UUUCCUUUCCACAC-CCCCCAAGAAGCAAAGAAAAACUUGGAAAGGUUGGCACAUUUUCCACCUCUCCCGGUCGAAGUUGGGGGCGAAAA-AA .............(-(((((((...((.......(((((....))))).))....((((.(((......))).))))))))))).)....-.. ( -22.40) >DroSec_CAF1 27955 72 + 1 UUCCCUUUC-----GCCCCCAAGAAGCAAAGAAAAACUUGGAAAGUU----------------UCUCCCUGUUGAAGUUGGGGACGAAAAAAA .....((((-----(.((((((..((((..((.(((((.....))))----------------).))..))))....)))))).))))).... ( -21.10) >DroSim_CAF1 27939 91 + 1 UUCCCUUUCCAUACCCCCCCAAGAAGCAAAGAAAAACUUGGAAAGGUUGGCACAUUUUCCACCUCUCCCUGUCGAAGUUGGGGGCGAAA--AA .............(.(((((((......(((.....)))(((.((((.((........)))))).))).........))))))).)...--.. ( -21.10) >DroEre_CAF1 32054 88 + 1 UUCCCUUUCCACAC--CCCCAAGAAGCAUAGAAAAACUUGGAAAGGUUGGCACAUUUUCCACCUCUCCCUGUCGAAGUUGGGGG--AAAA-AA .............(--((((((...(.((((........(((.((((.((........)))))).))))))))....)))))))--....-.. ( -22.00) >DroYak_CAF1 28387 87 + 1 UUC-CUUUCCACAC--CCCCAAGAAGCAUAGAAAAACUUGGAAAGGUUGGUACAUUUUCUACCUCUCCCUUUCGAAGUUGGGUC--AAAA-AA ...-..........--..............((..(((((.((((((..((((.......))))....)))))).)))))...))--....-.. ( -18.30) >consensus UUCCCUUUCCACAC_CCCCCAAGAAGCAAAGAAAAACUUGGAAAGGUUGGCACAUUUUCCACCUCUCCCUGUCGAAGUUGGGGGCGAAAA_AA ................((((((.(((..........)))(((.(((.(((........)))))).))).........)))))).......... (-12.73 = -13.36 + 0.63)

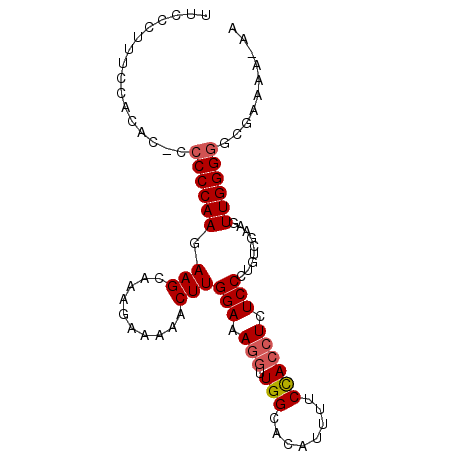

| Location | 13,662,903 – 13,662,994 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -19.09 |
| Energy contribution | -20.36 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13662903 91 - 27905053 UU-UUUUCGCCCCCAACUUCGACCGGGAGAGGUGGAAAAUGUGCCAACCUUUCCAAGUUUUUCUUUGCUUCUUGGGGG-GUGUGGAAAGGAAA ((-((..(((((((...........((((((((((........)).))))))))((((........))))....))))-)))..))))..... ( -32.30) >DroSec_CAF1 27955 72 - 1 UUUUUUUCGUCCCCAACUUCAACAGGGAGA----------------AACUUUCCAAGUUUUUCUUUGCUUCUUGGGGGC-----GAAAGGGAA ((((((((((((((((......((((((((----------------((((.....))))))))))))....))))))))-----)))))))). ( -32.90) >DroSim_CAF1 27939 91 - 1 UU--UUUCGCCCCCAACUUCGACAGGGAGAGGUGGAAAAUGUGCCAACCUUUCCAAGUUUUUCUUUGCUUCUUGGGGGGGUAUGGAAAGGGAA ((--((((((((((...........((((((((((........)).))))))))((((........)))).....))))))..)))))).... ( -29.80) >DroEre_CAF1 32054 88 - 1 UU-UUUU--CCCCCAACUUCGACAGGGAGAGGUGGAAAAUGUGCCAACCUUUCCAAGUUUUUCUAUGCUUCUUGGGG--GUGUGGAAAGGGAA ..-..((--(((........(((..((((((((((........)).))))))))..)))(((((((((((.....))--)))))))))))))) ( -31.70) >DroYak_CAF1 28387 87 - 1 UU-UUUU--GACCCAACUUCGAAAGGGAGAGGUAGAAAAUGUACCAACCUUUCCAAGUUUUUCUAUGCUUCUUGGGG--GUGUGGAAAG-GAA ..-....--...(((((((.((((((....((((.......))))..)))))).)))))(((((((((((.....))--))))))))))-).. ( -25.80) >consensus UU_UUUUCGCCCCCAACUUCGACAGGGAGAGGUGGAAAAUGUGCCAACCUUUCCAAGUUUUUCUUUGCUUCUUGGGGG_GUGUGGAAAGGGAA .........(((((((.........((((((((((........))).)))))))((((........)))).)))))))............... (-19.09 = -20.36 + 1.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:21 2006