
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,660,322 – 13,660,532 |
| Length | 210 |
| Max. P | 0.996899 |

| Location | 13,660,322 – 13,660,426 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.29 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -27.27 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
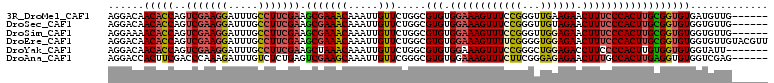
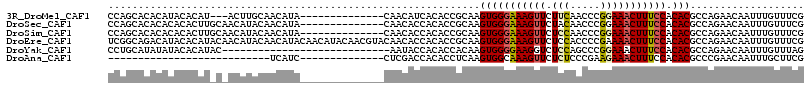
>3R_DroMel_CAF1 13660322 104 + 27905053 AGGACAACACCAGUCGAAGGAUUUGCCUUCGAAGCGAAACAAAUUGUUCUGGCGUGUGGAAAGUUUCCGGGUUGAAGAACUUUCCCACUUGCGGUGUGAUGUUG------ ..(((((((((..(((((((.....))))))).(((((((.....))).....(((.(((((((((..........)))))))))))))))))))))..)))).------ ( -35.30) >DroSec_CAF1 25489 104 + 1 AGGACAACACCAGUCGAAGGAUUUGCCUUCGAAGCGAAACAAAUUGUUCUGGCGUGUGGAAAGUUUCCGGGUUGUAGAACUUUCCCACUUGCGGUGUGGUGUUG------ ....((((((((.(((((((.....))))))).(((((((.....))).....(((.(((((((((..........))))))))))))))))....))))))))------ ( -37.00) >DroSim_CAF1 25355 104 + 1 AGGAAAACACCAGUCGAAGGAUUUGCCUUCGAAGCGAAACAAAUUGUUCUGGCGUGUGGAAAGUUUCCGGGUUGGAGAACUUUCCCACUUGCGGUGUGGUGUUG------ .....(((((((.(((((((.....))))))).(((((((.....))).....(((.(((((((((((.....)))).))))))))))))))....))))))).------ ( -38.30) >DroEre_CAF1 29446 110 + 1 AGGACAACACCAGUCGAAGGAUUUGCCUUCGAAGCGAAACAAAUUGUUCUGGCGUGUGGAAAGUUUUCGGGGUGGAGAACUUUCCCACUUGCGGUGUGGUGUUGUACGUU .(.(((((((((.(((((((.....))))))).(((((((.....))).....(((.(((((((((((......))))))))))))))))))....))))))))).)... ( -45.00) >DroYak_CAF1 25877 103 + 1 AGGACAACACCAGUCGAAGGAUUUGCCUUCGAAGCUAAACAAAUUGUUCUGGCGUGUGGAAAGUUUCCGGGCUGGAGACCUUCCCCACUUGUGGUGUGGUAUU------- ...((.((((((.(((((((.....))))))).(((((((.....))).))))(((.((((.((((((.....)))))).)))).)))...)))))).))...------- ( -39.70) >DroAna_CAF1 21116 104 + 1 AGGACCACUUCGACCCAAAGAUUUGUCUCUGAGUCGAAGCAAAUUGUUCGGGCGUGUGGAAAGUUUCUUCGGGAGAGAACUUUGCCACUUGAGGUGUGGUCGAG------ ..((((((.(((.(((...((((((..((......))..))))))....))))..(((((((((((..((....))))))))).))))....)).))))))...------ ( -35.80) >consensus AGGACAACACCAGUCGAAGGAUUUGCCUUCGAAGCGAAACAAAUUGUUCUGGCGUGUGGAAAGUUUCCGGGUUGGAGAACUUUCCCACUUGCGGUGUGGUGUUG______ ......(((((..(((((((.....))))))).(((((((.....))).....(((.((((((((((((...)))))).))))))))))))))))))............. (-27.27 = -27.50 + 0.23)
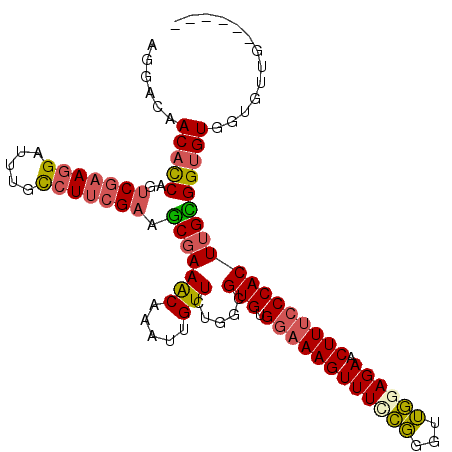
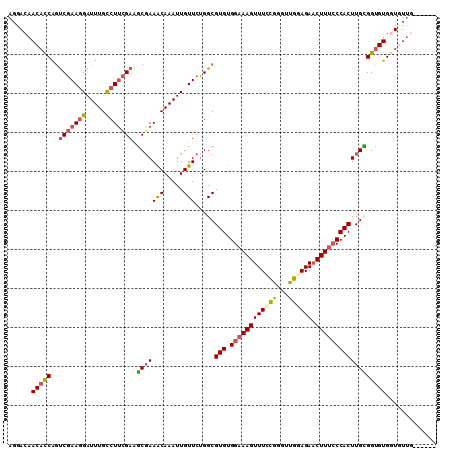
| Location | 13,660,322 – 13,660,426 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.29 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.29 |
| Mean z-score | -4.84 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
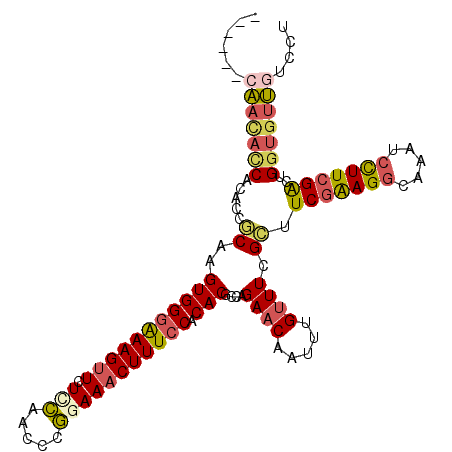
>3R_DroMel_CAF1 13660322 104 - 27905053 ------CAACAUCACACCGCAAGUGGGAAAGUUCUUCAACCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUCGCUUCGAAGGCAAAUCCUUCGACUGGUGUUGUCCU ------((((((((....((..(((((((((((.(((.....))))))))))).)))....((((.....)))).)).(((((((.....))))))).)))))))).... ( -34.00) >DroSec_CAF1 25489 104 - 1 ------CAACACCACACCGCAAGUGGGAAAGUUCUACAACCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUCGCUUCGAAGGCAAAUCCUUCGACUGGUGUUGUCCU ------((((((((....((..(((((((((((((.......)).)))))))).)))....((((.....)))).)).(((((((.....))))))).)))))))).... ( -35.90) >DroSim_CAF1 25355 104 - 1 ------CAACACCACACCGCAAGUGGGAAAGUUCUCCAACCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUCGCUUCGAAGGCAAAUCCUUCGACUGGUGUUUUCCU ------.(((((((....((..(((((((((((.(((.....))))))))))).)))....((((.....)))).)).(((((((.....))))))).)))))))..... ( -36.70) >DroEre_CAF1 29446 110 - 1 AACGUACAACACCACACCGCAAGUGGGAAAGUUCUCCACCCCGAAAACUUUCCACACGCCAGAACAAUUUGUUUCGCUUCGAAGGCAAAUCCUUCGACUGGUGUUGUCCU ...(.(((((((((....((..(((((((((((.((......)).)))))))).)))....((((.....)))).)).(((((((.....))))))).))))))))).). ( -39.00) >DroYak_CAF1 25877 103 - 1 -------AAUACCACACCACAAGUGGGGAAGGUCUCCAGCCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUAGCUUCGAAGGCAAAUCCUUCGACUGGUGUUGUCCU -------....((((.......))))(((((((.(((.....))).)))))))((((((((((((.....))).....(((((((.....))))))))))))).)))... ( -37.00) >DroAna_CAF1 21116 104 - 1 ------CUCGACCACACCUCAAGUGGCAAAGUUCUCUCCCGAAGAAACUUUCCACACGCCCGAACAAUUUGCUUCGACUCAGAGACAAAUCUUUGGGUCGAAGUGGUCCU ------...((((.........((((.((((((.(((.....)))))))))))))...............(((((((((((((((....))))))))))))))))))).. ( -39.30) >consensus ______CAACACCACACCGCAAGUGGGAAAGUUCUCCAACCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUCGCUUCGAAGGCAAAUCCUUCGACUGGUGUUGUCCU ......(((((((.....((..(((((((((((.(((.....))))))))))).)))....((((.....)))).)).(((((((.....)))))))..))))))).... (-26.98 = -27.68 + 0.70)

| Location | 13,660,356 – 13,660,455 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -19.40 |
| Consensus MFE | -12.58 |
| Energy contribution | -12.75 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13660356 99 - 27905053 CCAGCACACAUACACAU---ACUUGCAACAUA--------------CAACAUCACACCGCAAGUGGGAAAGUUCUUCAACCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUCG ...((...........(---((((((......--------------............)))))))((((((((.(((.....)))))))))))....))..((((.....)))).. ( -19.17) >DroSec_CAF1 25523 102 - 1 CCAGCACACACACACUUGCAACAUACAACAUA--------------CAACACCACACCGCAAGUGGGAAAGUUCUACAACCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUCG ...((.......(((((((.............--------------............)))))))((((((((((.......)).))))))))....))..((((.....)))).. ( -20.41) >DroSim_CAF1 25389 102 - 1 CCAGCACACACACACUUGCAACAUACAACAUA--------------CAACACCACACCGCAAGUGGGAAAGUUCUCCAACCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUCG ...((.......(((((((.............--------------............)))))))((((((((.(((.....)))))))))))....))..((((.....)))).. ( -24.01) >DroEre_CAF1 29480 116 - 1 UCGGCAGACAUACACAUACAACAUACAACAUACAACAUACAACGUACAACACCACACCGCAAGUGGGAAAGUUCUCCACCCCGAAAACUUUCCACACGCCAGAACAAUUUGUUUCG ..(((......................................((........)).......(((((((((((.((......)).)))))))).)))))).((((.....)))).. ( -20.10) >DroYak_CAF1 25911 88 - 1 CCUGCAUAUAUACACAUAC----------------------------AAUACCACACCACAAGUGGGGAAGGUCUCCAGCCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUAG ...((..............----------------------------....((((.......))))(((((((.(((.....))).)))))))....))..((((.....)))).. ( -20.80) >DroAna_CAF1 21150 75 - 1 ---------------------------UCAUC--------------CUCGACCACACCUCAAGUGGCAAAGUUCUCUCCCGAAGAAACUUUCCACACGCCCGAACAAUUUGCUUCG ---------------------------.....--------------................((((.((((((.(((.....))))))))))))).....((((........)))) ( -11.90) >consensus CCAGCACACAUACACAU_C_ACAUACAACAUA______________CAACACCACACCGCAAGUGGGAAAGUUCUCCAACCCGGAAACUUUCCACACGCCAGAACAAUUUGUUUCG ..............................................................(((((((((((.(((.....))))))))))).)))................... (-12.58 = -12.75 + 0.17)


| Location | 13,660,426 – 13,660,532 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -20.06 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13660426 106 + 27905053 --------UAUGUUGCAAGU---AUGUGUAUGUGUGCUGGAUGUGCUGGGUUGGGAAGGUGGGUCCUUCAGCUGGCUGUUCCAGUGUGACAAGCCACUGAGGUGGCAUAUUUGACAA --------....((((((((---((.....((((..(((((...((..(.....(((((.....)))))..)..))...)))))..).))).(((((....)))))))))))).))) ( -38.50) >DroSec_CAF1 25593 109 + 1 --------UAUGUUGUAUGUUGCAAGUGUGUGUGUGCUGGAUGUGCUGGGUUGGGUAGGUGGGUCCUUCAGCUGGCUGUUCCAGUGUGACAAGCCACUGAGGUGGCAUAUUUGACAA --------..((((((((((..(.((((..((((..(((((...((..(.((((..(((.....))))))))..))...)))))..).)))...))))...)..))))))..)))). ( -39.60) >DroSim_CAF1 25459 109 + 1 --------UAUGUUGUAUGUUGCAAGUGUGUGUGUGCUGGAUGUGCUGGGUUGGGGAGGUGGGUCCUUCAGCUGGCUGUUCCAGUGUGACAAGCCACUGAGGUGGCAUAUUUGACAA --------..((((((((((..(.((((..((((..(((((...((..(.(..((((......))))..).)..))...)))))..).)))...))))...)..))))))..)))). ( -41.80) >DroEre_CAF1 29556 116 + 1 GUAUGUUGUAUGUUGUAUGUUGUAUGUGUAUGUCUGCCGAAUGUGCUGUGU-GCGGAUGUAGGUGCUUCAGCUGGCGGUUCCAGUGUGACAAGCCUCUGAGGUGCCAUAUUUGACAA ...(((((((..(.(((((((...((.(((....)))))))))))).)..)-))((((((.((..((((((..(((...((......))...))).))))))..)))))))))))). ( -33.00) >DroYak_CAF1 25980 95 + 1 ---------------------GUAUGUGUAUAUAUGCAGGAUAGGCUGGAU-GAGGAUGUGGGUGCUUUAGCUGGCGGUUCCGAUGUGUCAAGCCUCUGAGGUGGCAUAUUUGACAA ---------------------.....(((..((((((.....(((((.(((-(.((((((.(((......))).)))..)))....)))).)))))........))))))...))). ( -25.72) >consensus ________UAUGUUGUAUGUUGCAUGUGUAUGUGUGCUGGAUGUGCUGGGUUGGGGAGGUGGGUCCUUCAGCUGGCUGUUCCAGUGUGACAAGCCACUGAGGUGGCAUAUUUGACAA ..........................(((..(((((((...((..(((((.......(((.(((......))).)))..)))))..))....(((.....))))))))))...))). (-20.06 = -19.70 + -0.36)

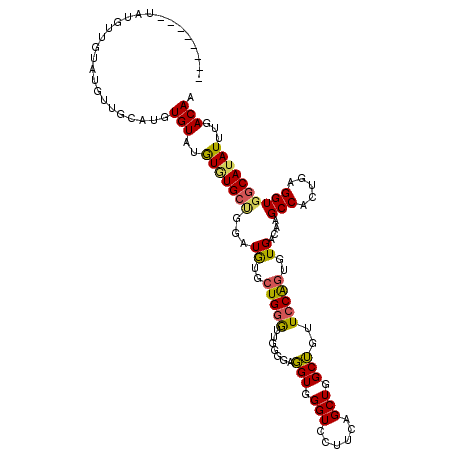

| Location | 13,660,426 – 13,660,532 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -19.45 |
| Consensus MFE | -9.78 |
| Energy contribution | -10.30 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13660426 106 - 27905053 UUGUCAAAUAUGCCACCUCAGUGGCUUGUCACACUGGAACAGCCAGCUGAAGGACCCACCUUCCCAACCCAGCACAUCCAGCACACAUACACAU---ACUUGCAACAUA-------- (((.(((.((((((((....))))).(((....(((((...((.....(((((.....)))))........))...)))))...))).....))---).))))))....-------- ( -21.52) >DroSec_CAF1 25593 109 - 1 UUGUCAAAUAUGCCACCUCAGUGGCUUGUCACACUGGAACAGCCAGCUGAAGGACCCACCUACCCAACCCAGCACAUCCAGCACACACACACUUGCAACAUACAACAUA-------- (((.(((....(((((....))))).(((....(((((.(((....))).(((.....)))...............)))))...))).....))))))...........-------- ( -20.00) >DroSim_CAF1 25459 109 - 1 UUGUCAAAUAUGCCACCUCAGUGGCUUGUCACACUGGAACAGCCAGCUGAAGGACCCACCUCCCCAACCCAGCACAUCCAGCACACACACACUUGCAACAUACAACAUA-------- (((.(((....(((((....))))).(((....(((((.(((....)))..(((......))).............)))))...))).....))))))...........-------- ( -20.10) >DroEre_CAF1 29556 116 - 1 UUGUCAAAUAUGGCACCUCAGAGGCUUGUCACACUGGAACCGCCAGCUGAAGCACCUACAUCCGC-ACACAGCACAUUCGGCAGACAUACACAUACAACAUACAACAUACAACAUAC .((((.....((((...((((.(........).))))....))))(((((((......)....((-.....))...)))))).)))).............................. ( -19.40) >DroYak_CAF1 25980 95 - 1 UUGUCAAAUAUGCCACCUCAGAGGCUUGACACAUCGGAACCGCCAGCUAAAGCACCCACAUCCUC-AUCCAGCCUAUCCUGCAUAUAUACACAUAC--------------------- .(((...((((((........(((((.((......((.....)).((....))............-.)).))))).....))))))..))).....--------------------- ( -16.22) >consensus UUGUCAAAUAUGCCACCUCAGUGGCUUGUCACACUGGAACAGCCAGCUGAAGGACCCACCUCCCCAACCCAGCACAUCCAGCACACAUACACAUACAACAUACAACAUA________ .((.(((....(((((....)))))))).))..(((((.......((((..((..........))....))))...))))).................................... ( -9.78 = -10.30 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:19 2006