
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,654,739 – 13,654,866 |
| Length | 127 |
| Max. P | 0.982749 |

| Location | 13,654,739 – 13,654,834 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.33 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -9.19 |
| Energy contribution | -9.33 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13654739 95 + 27905053 UCGGG---GGCUU---CUGGCUUUAUCUCUGCUUACUUUGCCACCUU---GUGGGUGGGACACUCUC-ACAUUCACCGUUUCAU---CC-----UCCGACUUCUCCUCGGAGU ..(((---(((..---..(((.........)))......))).))).---(((.(((((.....)))-))...)))........---.(-----(((((.......)))))). ( -25.50) >DroSec_CAF1 19752 98 + 1 UCGGG---GGCUU---CAGGCUUUAUCUCUGCUUACUUUGCCACCUCGUCGUGGGUGGCACACUCUC-ACAUUCUCCGUUUCAU---CC-----UCUGACUUCUCCUCGAAGU (((((---((...---.((((.........))))....((((((((......)))))))).......-................---))-----)))))((((.....)))). ( -26.50) >DroSim_CAF1 19587 98 + 1 UCGGG---GGCUU---CCGGCUUUAUCUCUGCUUACUUUGCCACCUCGUCGUGGGUGGCACACUCUC-ACAUUCUCCGCUUCAU---CC-----UCUGACUUCUCCUCGAAGU (((((---((...---..(((.........))).....((((((((......)))))))).......-................---))-----)))))((((.....)))). ( -25.30) >DroYak_CAF1 20095 98 + 1 UCGGG---GGUCA---CUGGCUUUAUCUCUGCUUACUUUGCCACCUU---AUGGGUGGGAACUACCA--GAUUUUCCUAUUC-U---CCUUUUCUCUGAAUUCUCUCUGGAGU .((((---(((..---........))))))).........(((((..---...)))))..(((.(((--((.......((((-.---..........))))....)))))))) ( -20.20) >DroAna_CAF1 15313 85 + 1 UCGGG---GGAAAAGUAUUUCUUUAUCUCUGCUUACUUUGCCACCUU---U---------CUCUCUC--CACUGGCAGUUCCAU-----------UUGCCUACUCCCCUGACU (((((---(((((((((................))))))........---.---------.......--....(((((......-----------)))))...)))))))).. ( -22.29) >DroPer_CAF1 25449 100 + 1 UCAGGUCGGAAAA---GUGUCUUUAUCUCUGCUUACUUUUCCACCUA---GCAGGGGGUUCCUCCCCUCCCCGCACCGC-CCAUGCCCCUACCCUCU--UUUCUCCCU----C ..((((.((((((---(((..............))))))))))))).---((.(((((........))))).)).....-.................--.........----. ( -24.84) >consensus UCGGG___GGCUA___CUGGCUUUAUCUCUGCUUACUUUGCCACCUU___GUGGGUGGCACACUCUC_ACAUUCUCCGUUUCAU___CC_____UCUGACUUCUCCCCGGAGU (((((...((((......))))..................((((((......))))))...............................................)))))... ( -9.19 = -9.33 + 0.14)



| Location | 13,654,766 – 13,654,866 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.17 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -11.56 |
| Energy contribution | -12.58 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13654766 100 + 27905053 UACUUUGCCACCUU---GUGGGUGGGACACUCUC-ACAUUCACCGUUUCAU---CC-----UCCGACUUCUCCUCGGAGUUCUCUAGUGCAGAUUGCC---CGAGUGGCCGUGUC----- .......(((((..---...)))))(((((....-.(((((..........---.(-----(((((.......)))))).......(.((.....)).---))))))...)))))----- ( -28.00) >DroSec_CAF1 19779 103 + 1 UACUUUGCCACCUCGUCGUGGGUGGCACACUCUC-ACAUUCUCCGUUUCAU---CC-----UCUGACUUCUCCUCGAAGUUCUCUAGUGCAGAUUGCC---CGAGUGGCCGUGUC----- (((..((((((((......))))))))(((((..-................---..-----...((((((.....)))))).....(.((.....)).---))))))...)))..----- ( -26.50) >DroSim_CAF1 19614 103 + 1 UACUUUGCCACCUCGUCGUGGGUGGCACACUCUC-ACAUUCUCCGCUUCAU---CC-----UCUGACUUCUCCUCGAAGUUCUCUAGUGCAGAUUGCC---CGAGUGGCCGUGUC----- (((..((((((((......))))))))(((((..-.((.(((.((((....---..-----...((((((.....))))))....)))).))).))..---.)))))...)))..----- ( -31.12) >DroEre_CAF1 23581 98 + 1 UACUUUGCCACCUU---GUGGGUGGGAACCUCAC--GAUUCUCUCUUUC-U---CC-----UCUGACUUCUCUACGGAGUUCUCUAGUUCAAAUUGCC---CGAGUGGCCGUGUC----- ......(((((...---.((((((((((.(....--).)))))......-.---..-----...((...(((....)))...))...........)))---)).)))))......----- ( -22.80) >DroYak_CAF1 20122 103 + 1 UACUUUGCCACCUU---AUGGGUGGGAACUACCA--GAUUUUCCUAUUC-U---CCUUUUCUCUGAAUUCUCUCUGGAGUUCUUUAGUUCAAAUUGCC---CUAGUGGCCAUGUC----- ......(((((...---..(((((((((((.(((--((.......((((-.---..........))))....))))))))))))...........)))---)..)))))......----- ( -25.60) >DroPer_CAF1 25479 110 + 1 UACUUUUCCACCUA---GCAGGGGGUUCCUCCCCUCCCCGCACCGC-CCAUGCCCCUACCCUCU--UUUCUCCCU----CUUUUAAUUUCAGAUUGCCUCUUGAGUGGCUCUGGCUGGGA ..........((((---((((((((..............(((....-...)))...........--...))))))----..........((((..(((........))))))))))))). ( -27.39) >consensus UACUUUGCCACCUU___GUGGGUGGGACACUCUC_ACAUUCUCCGUUUCAU___CC_____UCUGACUUCUCCUCGGAGUUCUCUAGUGCAGAUUGCC___CGAGUGGCCGUGUC_____ .......((((((......)))))).......................................((((((.....))))))..............(((........)))........... (-11.56 = -12.58 + 1.03)

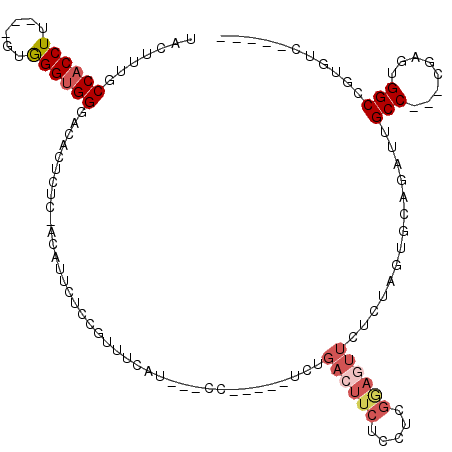
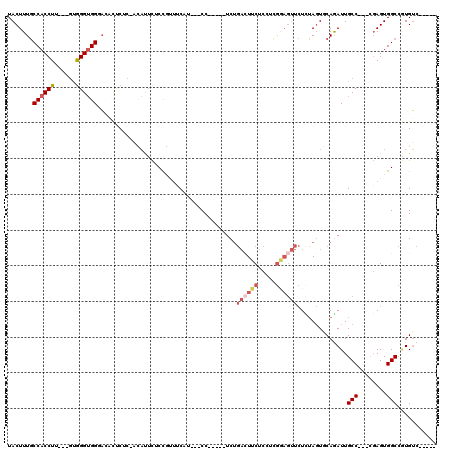
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:12 2006