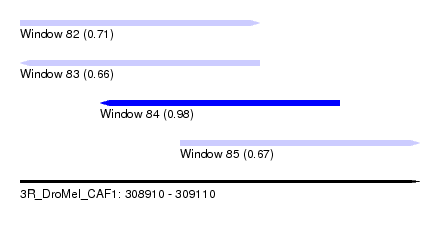
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 308,910 – 309,110 |
| Length | 200 |
| Max. P | 0.982920 |

| Location | 308,910 – 309,030 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -42.58 |
| Consensus MFE | -34.70 |
| Energy contribution | -35.30 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 308910 120 + 27905053 AGUGACCACCAGUUCGCCAUGAAGUGGAACGUGUACGCAUGGCCGCACCAGAUAAUGGUGCGAUUAUCGACAUUGAAGGACGAGCAGAAGGUGGACAUCGAGGACUUUCUCCGCCUGCAC .(((.(((((.(((((((.(.((((.((.((((....))))..(((((((.....)))))))....)).)).)).).)).)))))....))))).....((((....)))))))...... ( -39.30) >DroSec_CAF1 45312 120 + 1 AGUGACCACCAGUUCGCCAUGAAGUGGAACGUGUAUGCCUGGCCGCACCAGAUAAUGGUGCGGUUAUCCACCUUGAAAGACGAGCAGAAGGUGGACAUCGAGGACUUUCUCCGCCUGCAC ........(((.(((.....))).)))...(((((.((.(((((((((((.....)))))))))))((((((((.............))))))))......(((.....))))).))))) ( -43.92) >DroSim_CAF1 46159 120 + 1 AGUGACCACCAGUUCGCCAUGAAGUGGAACGUGUAUGCCUGGCCGCACCAGAUAAUGGUGCGGUUAUCCACCUUGAAAGACGAGCAGAAGGUGGACAUCGAGGACUUUCUCCGCCUGCAC ........(((.(((.....))).)))...(((((.((.(((((((((((.....)))))))))))((((((((.............))))))))......(((.....))))).))))) ( -43.92) >DroEre_CAF1 48232 120 + 1 AGUGACCACCAGUUCGCUAUGAAGUGGAACGUGUAUGCGUGGCCCCACCAGAUAAUGGUGCGAUUGUCCACCUUGAAAGACGAGCAGAAGGUGGACAUUGAGGAUUUUCUCCGCCUGCAC ......(((...((((((....))))))..)))..((((.(((..(((((.....)))))((((.(((((((((.............))))))))))))).(((.....)))))))))). ( -39.32) >DroYak_CAF1 48048 120 + 1 AGUGACCACCAGUUCGCCAUGAAGUGGAACGUGUAUGCAUGGCCGCACCAGAUAAUGGUGCGAUUGUCCACCUUGAAAGACGAGCAGAAGGUGGACAUUGAGGAGUUUCUCCGCCUGCAC ........(((.(((.....))).)))........((((.((((((((((.....)))))))..((((((((((.............))))))))))....((((...))))))))))). ( -46.42) >consensus AGUGACCACCAGUUCGCCAUGAAGUGGAACGUGUAUGCAUGGCCGCACCAGAUAAUGGUGCGAUUAUCCACCUUGAAAGACGAGCAGAAGGUGGACAUCGAGGACUUUCUCCGCCUGCAC ........(((.(((.....))).)))...(((((.((.....(((((((.....)))))))....((((((((.............))))))))......(((.....))))).))))) (-34.70 = -35.30 + 0.60)

| Location | 308,910 – 309,030 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -35.98 |
| Energy contribution | -36.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 308910 120 - 27905053 GUGCAGGCGGAGAAAGUCCUCGAUGUCCACCUUCUGCUCGUCCUUCAAUGUCGAUAAUCGCACCAUUAUCUGGUGCGGCCAUGCGUACACGUUCCACUUCAUGGCGAACUGGUGGUCACU (.(((((.((.(...(((...)))...).)).))))).)(.((.(((...((......(((((((.....)))))))((((((((....))........))))))))..))).)).)... ( -34.40) >DroSec_CAF1 45312 120 - 1 GUGCAGGCGGAGAAAGUCCUCGAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGAUAACCGCACCAUUAUCUGGUGCGGCCAGGCAUACACGUUCCACUUCAUGGCGAACUGGUGGUCACU ((((.(((.(((......)))..((((((((((.............))))))))))..(((((((.....))))))))))..))))....((.(((((....(.....).)))))..)). ( -40.72) >DroSim_CAF1 46159 120 - 1 GUGCAGGCGGAGAAAGUCCUCGAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGAUAACCGCACCAUUAUCUGGUGCGGCCAGGCAUACACGUUCCACUUCAUGGCGAACUGGUGGUCACU ((((.(((.(((......)))..((((((((((.............))))))))))..(((((((.....))))))))))..))))....((.(((((....(.....).)))))..)). ( -40.72) >DroEre_CAF1 48232 120 - 1 GUGCAGGCGGAGAAAAUCCUCAAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGACAAUCGCACCAUUAUCUGGUGGGGCCACGCAUACACGUUCCACUUCAUAGCGAACUGGUGGUCACU ((((.((((((.....)))....((((((((((.............))))))))))..(.(((((.....))))).))))..))))....((.(((((............)))))..)). ( -38.02) >DroYak_CAF1 48048 120 - 1 GUGCAGGCGGAGAAACUCCUCAAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGACAAUCGCACCAUUAUCUGGUGCGGCCAUGCAUACACGUUCCACUUCAUGGCGAACUGGUGGUCACU ((((((((((((...))))....((((((((((.............))))))))))..(((((((.....)))))))))).)))))....((.(((((....(.....).)))))..)). ( -45.72) >consensus GUGCAGGCGGAGAAAGUCCUCGAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGAUAAUCGCACCAUUAUCUGGUGCGGCCAGGCAUACACGUUCCACUUCAUGGCGAACUGGUGGUCACU ((((.(((.(((......)))..((((((((((.............))))))))))..(((((((.....))))))))))..))))....((.(((((............)))))..)). (-35.98 = -36.18 + 0.20)

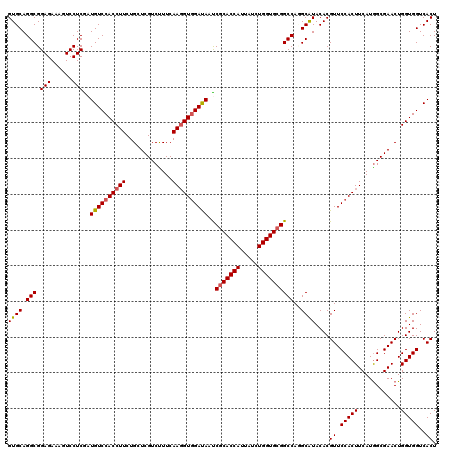
| Location | 308,950 – 309,070 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -46.20 |
| Consensus MFE | -40.50 |
| Energy contribution | -41.18 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 308950 120 - 27905053 GGGACUCCAGACGCUCCUCGAACGCUUGGCACUCGCUGGCGUGCAGGCGGAGAAAGUCCUCGAUGUCCACCUUCUGCUCGUCCUUCAAUGUCGAUAAUCGCACCAUUAUCUGGUGCGGCC .((((((..(((.((((((..(((((.(((....))))))))...)).))))...)))...)).)))).......(((((.(.......).)))....(((((((.....))))))))). ( -39.30) >DroSec_CAF1 45352 120 - 1 GGGACUCCAGUCGCUCCUCGAACGCUUGGCACUCGCUGGCGUGCAGGCGGAGAAAGUCCUCGAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGAUAACCGCACCAUUAUCUGGUGCGGCC ((((((....(((((......(((((.(((....))))))))...)))))....))))))...((((((((((.............)))))))))).((((((((.....)))))))).. ( -48.72) >DroSim_CAF1 46199 120 - 1 GGGACUCCAGUCGCUCCUCGAACGCUUGGCACUCGCUGGCGUGCAGGCGGAGAAAGUCCUCGAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGAUAACCGCACCAUUAUCUGGUGCGGCC ((((((....(((((......(((((.(((....))))))))...)))))....))))))...((((((((((.............)))))))))).((((((((.....)))))))).. ( -48.72) >DroEre_CAF1 48272 120 - 1 GCGACUCCAGACGCUCCUCAAACGCCUGGCACUCGCUGGAGUGCAGGCGGAGAAAAUCCUCAAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGACAAUCGCACCAUUAUCUGGUGGGGCC ((((((((.((......))....(((((.(((((....))))))))))))))...........((((((((((.............)))))))))).)))).(((((....))))).... ( -47.32) >DroYak_CAF1 48088 120 - 1 GGGACUCCAGACGCUCCUCAAACGCCUGGCACUCGCUGGCGUGCAGGCGGAGAAACUCCUCAAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGACAAUCGCACCAUUAUCUGGUGCGGCC ((((((((.((......))....(((((.(((.(....).))))))))))))....))))...((((((((((.............)))))))))).((((((((.....)))))))).. ( -46.92) >consensus GGGACUCCAGACGCUCCUCGAACGCUUGGCACUCGCUGGCGUGCAGGCGGAGAAAGUCCUCGAUGUCCACCUUCUGCUCGUCUUUCAAGGUGGAUAAUCGCACCAUUAUCUGGUGCGGCC ((((((.......((((((..(((((.(((....))))))))...)).))))..))))))...((((((((((.............)))))))))).((((((((.....)))))))).. (-40.50 = -41.18 + 0.68)

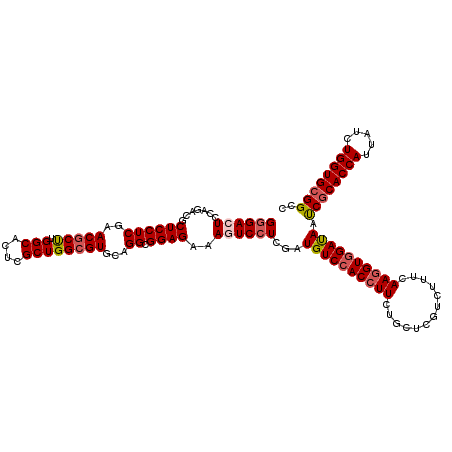
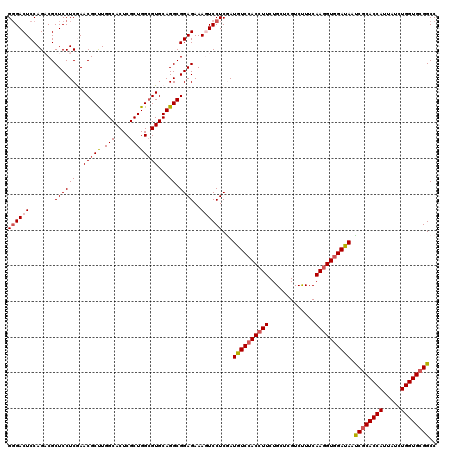
| Location | 308,990 – 309,110 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -47.21 |
| Consensus MFE | -42.24 |
| Energy contribution | -42.16 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 308990 120 + 27905053 CGAGCAGAAGGUGGACAUCGAGGACUUUCUCCGCCUGCACGCCAGCGAGUGCCAAGCGUUCGAGGAGCGUCUGGAGUCCCUCAACGACGAGGUGCAGGCCUACUCACUGGCGUUUAACAC ...(((...((((((..............)))))))))(((((((.((((((((..(((((...)))))..))).(..((((......))))..).....))))).)))))))....... ( -46.74) >DroSec_CAF1 45392 120 + 1 CGAGCAGAAGGUGGACAUCGAGGACUUUCUCCGCCUGCACGCCAGCGAGUGCCAAGCGUUCGAGGAGCGACUGGAGUCCCUCAACGACGAGGUGCAGGCCUACUCACUGGCGUUUAAUAC ...(((...((((((..............)))))))))(((((((.(((((((.(((((((...))))).))...(..((((......))))..).)))..)))).)))))))....... ( -45.24) >DroSim_CAF1 46239 120 + 1 CGAGCAGAAGGUGGACAUCGAGGACUUUCUCCGCCUGCACGCCAGCGAGUGCCAAGCGUUCGAGGAGCGACUGGAGUCCCUCAACGACGAGGUGCAGGCCUACUCACUGGCGUUUAAUAC ...(((...((((((..............)))))))))(((((((.(((((((.(((((((...))))).))...(..((((......))))..).)))..)))).)))))))....... ( -45.24) >DroEre_CAF1 48312 120 + 1 CGAGCAGAAGGUGGACAUUGAGGAUUUUCUCCGCCUGCACUCCAGCGAGUGCCAGGCGUUUGAGGAGCGUCUGGAGUCGCUCAACGACGAGGUGCAGGCCUACUCACUGGCGUUUAAUAC ((..(((..(((((.......(((.....)))((((((((..((((((.(.((((((((((...))))))))))).))))).......)..)))))))))))))..))).))........ ( -48.81) >DroYak_CAF1 48128 120 + 1 CGAGCAGAAGGUGGACAUUGAGGAGUUUCUCCGCCUGCACGCCAGCGAGUGCCAGGCGUUUGAGGAGCGUCUGGAGUCCCUCAACGACGAGGUGCAGGCCUACUCACUGGCGUUUAAUAC ...(((...((((((..............)))))))))(((((((.(((((((((((((((...)))))))))).(..((((......))))..).....))))).)))))))....... ( -50.04) >consensus CGAGCAGAAGGUGGACAUCGAGGACUUUCUCCGCCUGCACGCCAGCGAGUGCCAAGCGUUCGAGGAGCGUCUGGAGUCCCUCAACGACGAGGUGCAGGCCUACUCACUGGCGUUUAAUAC ...(((...((((((..............)))))))))(((((((.(((((((.(((((((...))))).))...((.((((......)))).)).)))..)))).)))))))....... (-42.24 = -42.16 + -0.08)

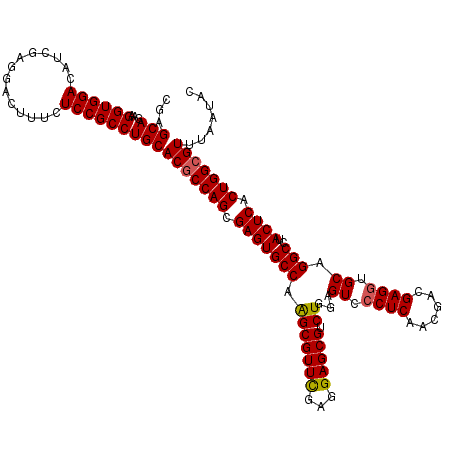
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:28 2006