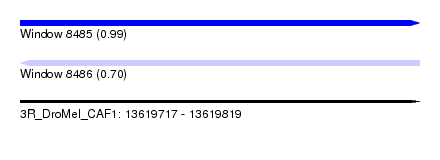
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,619,717 – 13,619,819 |
| Length | 102 |
| Max. P | 0.991693 |

| Location | 13,619,717 – 13,619,819 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -20.64 |
| Energy contribution | -22.46 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
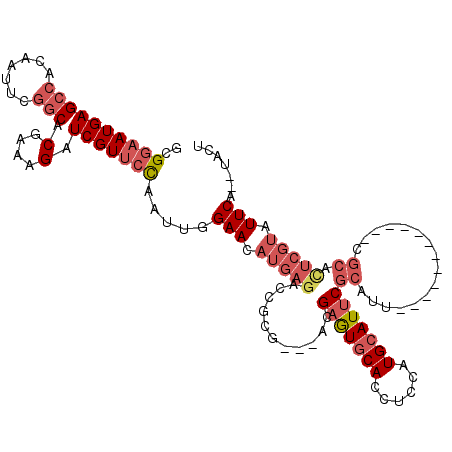
>3R_DroMel_CAF1 13619717 102 + 27905053 GCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCG---ACGAGUGCACCUCCAUGCAUUCGCAUU------------CGCACUCGUAUUCA--UACU ..((((((((((.......))).(....).))))))).....(((.(((((...(((---(((((((((......))))))))...)------------))).))))).))).--.... ( -36.70) >DroSec_CAF1 68684 102 + 1 GCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCG---ACGAGUGCACCUCCAUGCAUUCGCAUU------------CGCACUCGUAUUCA--UACU ..((((((((((.......))).(....).))))))).....(((.(((((...(((---(((((((((......))))))))...)------------))).))))).))).--.... ( -36.70) >DroSim_CAF1 62308 102 + 1 GCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCG---ACGAGUGCACCUCCAUGCAUUCGCAUU------------CGCACUCGUAUUCA--UACU ..((((((((((.......))).(....).))))))).....(((.(((((...(((---(((((((((......))))))))...)------------))).))))).))).--.... ( -36.70) >DroEre_CAF1 59961 113 + 1 GCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCG---ACGAGUGCACCUCCAUGCAUUCGCAUUCGCAU-CGCACUCGCAUUCGUAUUCA--UACU ..((((((((((.......))).(....).))))))).....(((.(((((...(((---(((((((((......))))))))...))))..-.((....)).))))).))).--.... ( -37.20) >DroYak_CAF1 61782 113 + 1 -CGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCG---AAGAGUGCACCUCCAUGCAUUCGCAAUCGCAUUUGCACUCGCACUCGUAUUCA--UACU -.((((((((((.......))).(....).))))))).....(((.(((((...(((---(.(((((((......)))))))((((......))))..)))).))))).))).--.... ( -39.80) >DroPer_CAF1 69877 88 + 1 AUGGUAUGAGGAAUGAUUGCGC-------UUCGUUAUAAAUGGAACGUGAGUCUACACAUUCGUAUUCACAUUCCUGCAUUCAUA------------------------UUCAUGUAUU ((((.((((((((((.(..((.-------(((((.....))))).))..)(..(((......)))..).))))))).))))))).------------------------.......... ( -19.90) >consensus GCGGAAUGAGCCACAAUUCGGCACGAAAGAUCGUUCCAAUUGGAACAUGAGACCGCG___ACGAGUGCACCUCCAUGCAUUCGCAUU____________CGCACUCGUAUUCA__UACU ..((((((((((.......))).(....).))))))).....(((.(((((...........(((((((......)))))))((................)).))))).)))....... (-20.64 = -22.46 + 1.82)

| Location | 13,619,717 – 13,619,819 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -18.24 |
| Energy contribution | -20.80 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
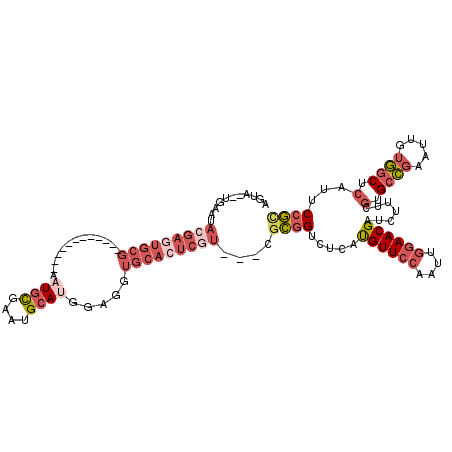
>3R_DroMel_CAF1 13619717 102 - 27905053 AGUA--UGAAUACGAGUGCG------------AAUGCGAAUGCAUGGAGGUGCACUCGU---CGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGC ....--.....(((((((((------------.((((....)))).....)))))))))---.((((......((..(((((((.((((....)))).)).)))))..)).....)))) ( -36.00) >DroSec_CAF1 68684 102 - 1 AGUA--UGAAUACGAGUGCG------------AAUGCGAAUGCAUGGAGGUGCACUCGU---CGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGC ....--.....(((((((((------------.((((....)))).....)))))))))---.((((......((..(((((((.((((....)))).)).)))))..)).....)))) ( -36.00) >DroSim_CAF1 62308 102 - 1 AGUA--UGAAUACGAGUGCG------------AAUGCGAAUGCAUGGAGGUGCACUCGU---CGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGC ....--.....(((((((((------------.((((....)))).....)))))))))---.((((......((..(((((((.((((....)))).)).)))))..)).....)))) ( -36.00) >DroEre_CAF1 59961 113 - 1 AGUA--UGAAUACGAAUGCGAGUGCG-AUGCGAAUGCGAAUGCAUGGAGGUGCACUCGU---CGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGC .(((--....)))....(((((((((-...(..((((....)))).)...)))))))))---.((((......((..(((((((.((((....)))).)).)))))..)).....)))) ( -37.50) >DroYak_CAF1 61782 113 - 1 AGUA--UGAAUACGAGUGCGAGUGCAAAUGCGAUUGCGAAUGCAUGGAGGUGCACUCUU---CGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCG- .(.(--(((.(((((...((.(..(..(((.((((((((((((((....)))))...))---))))))).)))(((((....)))))........)..)))..)))))..)))).)..- ( -37.40) >DroPer_CAF1 69877 88 - 1 AAUACAUGAA------------------------UAUGAAUGCAGGAAUGUGAAUACGAAUGUGUAGACUCACGUUCCAUUUAUAACGAA-------GCGCAAUCAUUCCUCAUACCAU .....((((.------------------------.((((.(((.(((((((((.((((....))))...))))))))).(((.....)))-------..))).))))...))))..... ( -22.00) >consensus AGUA__UGAAUACGAGUGCG____________AAUGCGAAUGCAUGGAGGUGCACUCGU___CGCGGUCUCAUGUUCCAAUUGGAACGAUCUUUCGUGCCGAAUUGUGGCUCAUUCCGC ...........(((((((((.............((((....)))).....)))))))))....((((.....((((((....)))))).......(.((((.....)))).)...)))) (-18.24 = -20.80 + 2.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:53 2006