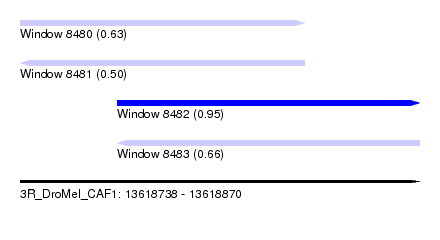
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,618,738 – 13,618,870 |
| Length | 132 |
| Max. P | 0.951728 |

| Location | 13,618,738 – 13,618,832 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -25.61 |
| Energy contribution | -26.95 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13618738 94 + 27905053 AUAUCCGCGAGACGUCUCGGGAUUGUCCCAUU-------GCGAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUUGCUCGGA--------G ...((((.(((....)))(((.....)))...-------((((((((....((((.(((((..((......))..)))))...)))))))))))).))))--------. ( -37.10) >DroSec_CAF1 67715 94 + 1 AUAUCCGCGAGACGUCUCGGGAUUGUCCCAUU-------GCGAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUCGCUCGGA--------G ...((((.(((....)))(((.....)))...-------((((((((....((((.(((((..((......))..)))))...)))))))))))).))))--------. ( -39.20) >DroSim_CAF1 61334 94 + 1 AUAUCCGCGAGACGUCUCGGGAUUGUCCCAUU-------GCGAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUCGCUCGGA--------G ...((((.(((....)))(((.....)))...-------((((((((....((((.(((((..((......))..)))))...)))))))))))).))))--------. ( -39.20) >DroEre_CAF1 58986 94 + 1 AUAUCCGCGAGACGUCUCGGGAUUGUCCCAUU-------GCGAAGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUUACUUGGA--------G ...((((((((....)))(((.....)))...-------)).((((.....((((.(((((..((......))..)))))...))))......)))))))--------. ( -33.70) >DroYak_CAF1 60331 102 + 1 AUAUCCGCGAGACGUCUCGGGAUUGUCCCAUU-------GCGAAGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCAGCUUAACUUGGAUGAACAGAG .((((((((((....)))(((.....)))...-------)).(((((.((.((((.(((((..((......))..)))))...))))))...))))))))))....... ( -35.70) >DroAna_CAF1 58210 89 + 1 AUAUCCCCAAGACGUCUCGGGAUUGUCUCUUUUUUUUGUGCACUGGGCCUAGAUCGAGUUGGCUCGAUAAAGGUGCAGCUGCAGU------------GGA--------A ...(((.(.(((((((....)).)))))..........(((((((.((((..((((((....))))))..)))).))).))))).------------)))--------. ( -32.20) >consensus AUAUCCGCGAGACGUCUCGGGAUUGUCCCAUU_______GCGAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUCGCUCGGA________G ...((((((((....)))(((.....)))..........))((((((....((((.(((((..((......))..)))))...))))))))))....)))......... (-25.61 = -26.95 + 1.34)



| Location | 13,618,738 – 13,618,832 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -21.49 |
| Energy contribution | -21.38 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13618738 94 - 27905053 C--------UCCGAGCAAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAU .--------..((((....((((((.(((....))).....))))))....))))...........((((-------...(((.....)))(((....))))))).... ( -27.80) >DroSec_CAF1 67715 94 - 1 C--------UCCGAGCGAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAU .--------..((((....((((((.(((....))).....))))))....))))...........((((-------...(((.....)))(((....))))))).... ( -27.80) >DroSim_CAF1 61334 94 - 1 C--------UCCGAGCGAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAU .--------..((((....((((((.(((....))).....))))))....))))...........((((-------...(((.....)))(((....))))))).... ( -27.80) >DroEre_CAF1 58986 94 - 1 C--------UCCAAGUAAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACUUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAU .--------(((.......((((((.(((....))).....)))))).(.....).....)))..(((((-------...(((.....)))(((....))))))))... ( -25.70) >DroYak_CAF1 60331 102 - 1 CUCUGUUCAUCCAAGUUAAGCUGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACUUCGC-------AAUGGGACAAUCCCGAGACGUCUCGCGGAUAU ........(((((((((.(((((.....)))))...(((..(((((......)))))..)))))))).((-------...(((.....)))(((....))))))))).. ( -30.80) >DroAna_CAF1 58210 89 - 1 U--------UCC------------ACUGCAGCUGCACCUUUAUCGAGCCAACUCGAUCUAGGCCCAGUGCACAAAAAAAAGAGACAAUCCCGAGACGUCUUGGGGAUAU .--------...------------..((((.(((..(((..((((((....))))))..)))..)))))))................(((((((....))))))).... ( -29.00) >consensus C________UCCGAGCAAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC_______AAUGGGACAAUCCCGAGACGUCUCGCGGAUAU .........(((..............(((....)))(((..(((((......)))))..)))...................)))..((((((((....)))).)))).. (-21.49 = -21.38 + -0.11)



| Location | 13,618,770 – 13,618,870 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -26.64 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13618770 100 + 27905053 GCGAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUUGCUCGGA--------G----GAGCUGAGGAGCA-AGGAGCAGGCGAGGAUAACGAUGGG ((...((((((.((((.(((((..((......))..)))))...)))).(((((((((...--------.----))))...))))).-))))))..))............... ( -37.70) >DroSec_CAF1 67747 100 + 1 GCGAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUCGCUCGGA--------G----GAGCAGAGGAGCA-AGGAGCAGGCGAGGAUCACGAUGGG ((...((((((.((((.(((((..((......))..)))))...)))).(((((((((...--------.----))))...))))).-))))))..))............... ( -39.20) >DroSim_CAF1 61366 100 + 1 GCGAGGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUCGCUCGGA--------G----GAGCAGAGGAGCA-AGGAGCAGGCGAGGAUCACGAUGGG ((...((((((.((((.(((((..((......))..)))))...)))).(((((((((...--------.----))))...))))).-))))))..))............... ( -39.20) >DroEre_CAF1 59018 101 + 1 GCGAAGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUUACUUGGA--------G----GAGCAGAGGAGCAGAAGAGCAGGCGAAGAUCAGGAUGGG ((...((((((.((((.(((((..((......))..)))))...)))).(((((.......--------)----))))..))))))......))................... ( -30.60) >DroYak_CAF1 60363 109 + 1 GCGAAGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCAGCUUAACUUGGAUGAACAGAG----AAGCAGAGGAGCAGAGUAGCAGGCGAGGAUCAGGAUGGG ((...((((((.((((.(((((..((......))..)))))...)))).((((...(((......)))..----))))..))))))......))................... ( -32.40) >DroPer_CAF1 68609 90 + 1 -UGGAGCGCCCAGAUCCAGUGUGAGCGAUAAAGGUGCAGCUGCUG----GCU--GCCCAGA--------GAAUGGGGCGGGGGGGCA-GGGGGCAGGCAGGCAGCA------- -....(((((...((((.......).)))...))))).(((((((----.((--((((...--------.......((......)).-..)))))).)).))))).------- ( -35.12) >consensus GCGAAGUUCCUAGAUCGAGUUGUGCCGAUAAAGGUGCAGCUGCAGAUCGGCUUCGCUCGGA________G____GAGCAGAGGAGCA_AGGAGCAGGCGAGGAUCACGAUGGG .((..((((((.((((.(((((((((......)))))))))...)))).(((((((((................))))...)))))..))))))...)).............. (-26.64 = -27.42 + 0.78)


| Location | 13,618,770 – 13,618,870 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -16.17 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13618770 100 - 27905053 CCCAUCGUUAUCCUCGCCUGCUCCU-UGCUCCUCAGCUC----C--------UCCGAGCAAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC ..........((((....(((((..-.(((....)))..----.--------...)))))..((((((.(((....))).....))))))............))))....... ( -21.60) >DroSec_CAF1 67747 100 - 1 CCCAUCGUGAUCCUCGCCUGCUCCU-UGCUCCUCUGCUC----C--------UCCGAGCGAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC ......((((.....((((((....-.(((..((.((((----.--------...)))))))))......))))..)).(((..(((((......)))))..)))....)))) ( -23.50) >DroSim_CAF1 61366 100 - 1 CCCAUCGUGAUCCUCGCCUGCUCCU-UGCUCCUCUGCUC----C--------UCCGAGCGAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC ......((((.....((((((....-.(((..((.((((----.--------...)))))))))......))))..)).(((..(((((......)))))..)))....)))) ( -23.50) >DroEre_CAF1 59018 101 - 1 CCCAUCCUGAUCUUCGCCUGCUCUUCUGCUCCUCUGCUC----C--------UCCAAGUAAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACUUCGC ....((((((((......((((.....((......))..----.--------....))))..((((((.(((....))).....)))))).......)))).))))....... ( -22.60) >DroYak_CAF1 60363 109 - 1 CCCAUCCUGAUCCUCGCCUGCUACUCUGCUCCUCUGCUU----CUCUGUUCAUCCAAGUUAAGCUGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACUUCGC ....((((((((...((((((....((((...((.((((----..((.........))..)))).))...))))..)))........))).......)))).))))....... ( -23.10) >DroPer_CAF1 68609 90 - 1 -------UGCUGCCUGCCUGCCCCC-UGCCCCCCCGCCCCAUUC--------UCUGGGC--AGC----CAGCAGCUGCACCUUUAUCGCUCACACUGGAUCUGGGCGCUCCA- -------.(((((.((.((((((..-..................--------...))))--)).----))))))).((.(((..(((.(.......))))..))).))....- ( -27.70) >consensus CCCAUCGUGAUCCUCGCCUGCUCCU_UGCUCCUCUGCUC____C________UCCGAGCAAAGCCGAUCUGCAGCUGCACCUUUAUCGGCACAACUCGAUCUAGGAACCUCGC ...............((((((......(((.....((((................))))..)))......))))..)).(((..(((((......)))))..)))........ (-16.17 = -15.97 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:50 2006