
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,606,228 – 13,606,338 |
| Length | 110 |
| Max. P | 0.780705 |

| Location | 13,606,228 – 13,606,338 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -56.23 |
| Consensus MFE | -41.19 |
| Energy contribution | -42.13 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
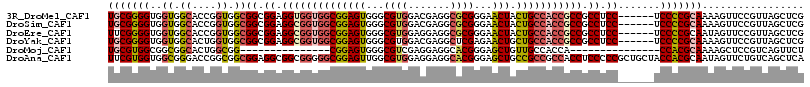
>3R_DroMel_CAF1 13606228 110 + 27905053 UGCGGGGUGGUGGCACCGGUGGCGGCGGAGGUGGUGGCGGAGUGGGCGUGGACGAGGCGCGGGAACUACUGCCACCGCCGCCUCC------UCCCCGCAAAAGUUCCGUUAGCUCG (((((((.((.(((.(((.......))).((((((((((((((...((((.......))))...))).)))))))))))))).))------.)))))))................. ( -58.90) >DroSim_CAF1 48899 110 + 1 UGCGGGGUGGUGGCACCGGUGGCGGCGGAGGCGGUGGCGGAGUGGGCGUGGACGAGGCGCGGGAACUACUGCCACCGCCGCCUCC------UCCCCGCAAAAGUUCCGUUAGCUCG (((((((.((.(((.(((.......))).((((((((((((((...((((.......))))...))).)))))))))))))).))------.)))))))................. ( -60.80) >DroEre_CAF1 46327 110 + 1 UUCGGGGUGGUGGCACCGGUGGCGGCGGAGGCGGUGGCGGAGUGGGCGUGGAGGAGGCGCGGGAACUACUGCCACCGCCGCCUCC------UCCCCGCAAUAGUUCCGUUAGCUCG ..((((.(..(((.((..((((.((.(((((((((((.(((((((.((((.......))))....))))).)).)))))))))))------.))))))....)).)))..).)))) ( -58.90) >DroYak_CAF1 47589 110 + 1 UGCGGGGUGGUGGCACUGGUGGCGGCGGAGGCGGUGGCGGAGUGGGCGUGGACGAGGCUCGAGAACUGCUGCCACCGCCGCCUCC------UCCCCGCAAAAGUUCCGUUAGCUCG ..((((.(..(((.(((.((((.((.(((((((((((.((((..(.((.(.......).))....)..)).)).)))))))))))------.))))))...))).)))..).)))) ( -58.40) >DroMoj_CAF1 17285 86 + 1 UGCGUGGCGGCGGCACUGGCGG---------------CGGAGUGGGCGUCGAGGAGGCACGGGAGCUGUUGCCACCA---------------CCACGCAAAAGCUCCGUCAGUUCU .((.((((.((.((....)).)---------------)(((((..((((.(.((.(((((((...))).)))).)).---------------).))))....)))))))))))... ( -35.00) >DroAna_CAF1 46047 116 + 1 UUCGUGGUGGCGGGACCGGCGGCGGAGGCGGCGGGGGCGGAGUUGGCGUGGAGGAGGCACGGGAGCUGCCGCCGCCACCUCCCCCGCUGCUACCACGCAAUAGUUCUGUCAGCUCA ...(.(.(((((((((..(((..((.(((((((((((.((...((((((((....(((......))).))).))))))).))))))))))).)).)))....))))))))).).). ( -65.40) >consensus UGCGGGGUGGUGGCACCGGUGGCGGCGGAGGCGGUGGCGGAGUGGGCGUGGACGAGGCGCGGGAACUACUGCCACCGCCGCCUCC______UCCCCGCAAAAGUUCCGUUAGCUCG (((((((..((.((....)).))((.((.((((((((((((((...((((.......))))...))).))))))))))).)).)).......)))))))................. (-41.19 = -42.13 + 0.95)


| Location | 13,606,228 – 13,606,338 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -46.80 |
| Consensus MFE | -30.31 |
| Energy contribution | -31.38 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
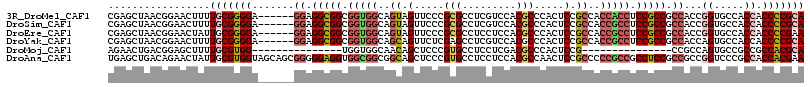
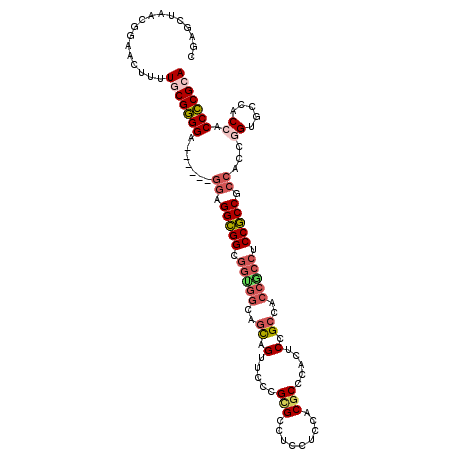
>3R_DroMel_CAF1 13606228 110 - 27905053 CGAGCUAACGGAACUUUUGCGGGGA------GGAGGCGGCGGUGGCAGUAGUUCCCGCGCCUCGUCCACGCCCACUCCGCCACCACCUCCGCCGCCACCGGUGCCACCACCCCGCA .................(((((((.------((.(((((.((((((.(.(((....(((.........)))..)))).)))))).))...((((....))))))).)).))))))) ( -46.40) >DroSim_CAF1 48899 110 - 1 CGAGCUAACGGAACUUUUGCGGGGA------GGAGGCGGCGGUGGCAGUAGUUCCCGCGCCUCGUCCACGCCCACUCCGCCACCGCCUCCGCCGCCACCGGUGCCACCACCCCGCA .................(((((((.------((.((((((((((((.(.(((....(((.........)))..)))).)))))))))...((((....))))))).)).))))))) ( -51.80) >DroEre_CAF1 46327 110 - 1 CGAGCUAACGGAACUAUUGCGGGGA------GGAGGCGGCGGUGGCAGUAGUUCCCGCGCCUCCUCCACGCCCACUCCGCCACCGCCUCCGCCGCCACCGGUGCCACCACCCCGAA ...................(((((.------((.((((((((((((.(.(((....(((.........)))..)))).)))))))))...((((....))))))).)).))))).. ( -47.90) >DroYak_CAF1 47589 110 - 1 CGAGCUAACGGAACUUUUGCGGGGA------GGAGGCGGCGGUGGCAGCAGUUCUCGAGCCUCGUCCACGCCCACUCCGCCACCGCCUCCGCCGCCACCAGUGCCACCACCCCGCA .................(((((((.------((((((((.(((((..((.((...((.....))...)))).....))))).))))))))(.(((.....))).)....))))))) ( -45.80) >DroMoj_CAF1 17285 86 - 1 AGAACUGACGGAGCUUUUGCGUGG---------------UGGUGGCAACAGCUCCCGUGCCUCCUCGACGCCCACUCCG---------------CCGCCAGUGCCGCCGCCACGCA .................(((((((---------------((((((((...((...((........))..((.......)---------------).))...))))))))))))))) ( -33.00) >DroAna_CAF1 46047 116 - 1 UGAGCUGACAGAACUAUUGCGUGGUAGCAGCGGGGGAGGUGGCGGCGGCAGCUCCCGUGCCUCCUCCACGCCAACUCCGCCCCCGCCGCCUCCGCCGCCGGUCCCGCCACCACGAA ...................((((((.((.((((.(((((((((((.(((((....((((.......))))....))..))).))))))))))).)))).(....))).)))))).. ( -55.90) >consensus CGAGCUAACGGAACUUUUGCGGGGA______GGAGGCGGCGGUGGCAGCAGUUCCCGCGCCUCCUCCACGCCCACUCCGCCACCGCCUCCGCCGCCACCGGUGCCACCACCCCGCA .................(((((((.......((.(((((.(((((..((.(.....(((.........))).....).))..))))).))))).))...((.....)).))))))) (-30.31 = -31.38 + 1.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:41 2006