| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,587,617 – 13,587,714 |
| Length | 97 |
| Max. P | 0.686449 |

| Location | 13,587,617 – 13,587,714 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 70.91 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -15.97 |
| Energy contribution | -18.08 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
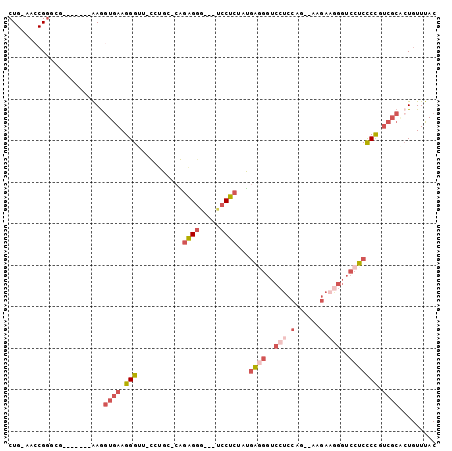
>3R_DroMel_CAF1 13587617 97 + 27905053 CUGGAACCGGGCGAUGGGUGAAGGUGAAGGGUUUUCUGC-CAGAGGG---UCCUCUUUGAGGGUCCUCCAG--AAGAAGGGUCCUCCCCGUCGCUAUGUUUAC ...((((..(((((((((.((.((.......(((((((.-..((((.---.((((...))))..)))))))--)))).....)))))))))))))..)))).. ( -43.80) >DroSec_CAF1 37110 91 + 1 CUGGAACCGGGCG-------AAGGUGAAGGGUUUUCAGCCCAGAGGG---UCCUCCAUGAGGGUCCUCCAG--AAGAUGGGUCCUCCCCGUCGCACUGUUUAC (((((...(((((-------((..(.....)..))).))))...((.---.((((...))))..)))))))--..((((((.....))))))........... ( -36.50) >DroSim_CAF1 29251 91 + 1 CUGGAACCGGGCG-------AAGGUGAAGGGUUUUCUGCCCAGAGGG---UCCUCUGUGAGGGUCCUCCAG--AAGAAGGGUCCUCCCCGUCGCGCUGUUUAC ...((((.(.(((-------(.((.(..((((.....)))).((((.---.((((((.(((....))))))--)....))..))))))).)))).).)))).. ( -39.20) >DroEre_CAF1 27764 93 + 1 CUG-AAGCGGCUG-------GAGGUGAAGGGUU-CCUGC-CGGAGGACCCUCCUCAACGAGGGUCCUCCAGAAGAGAAGGGUCCUCCCCGUCGCACUGUUUAC ...-..(((((.(-------((((.....(((.-...))-)(((((((((((......))))))))))).............)))))..)))))......... ( -43.70) >DroYak_CAF1 28086 88 + 1 CUG-AACCGGUUG-------AAGGUGAAGGGUU-CCUGA-CGGAGGG---UCCUCAAUGAUGGUCCUCCAG--GAGAAAGGUCCUCCCCGUCGCACUGUUUAC .((-(((.(((((-------(.((.(.((((((-(((..-.((((((---.((........))))))))))--))).....)))).))).))).)))))))). ( -26.90) >DroPer_CAF1 41109 73 + 1 CU------GCCAG-------GAACUAAAGGGU--CCUGA-AGAGGCC---UCCUCUCUCGUCUCCCCCCU-----------CUCUCUCUCCCUCUCUGUUUAC ..------.....-------((((...((((.--...((-(((((..---.................)))-----------)).))...))))....)))).. ( -12.61) >consensus CUG_AACCGGGCG_______AAGGUGAAGGGUU_CCUGC_CAGAGGG___UCCUCUAUGAGGGUCCUCCAG__AAGAAGGGUCCUCCCCGUCGCACUGUUUAC .......................((((.(((...........((((.....))))...((((..(((.(......).)))..))))))).))))......... (-15.97 = -18.08 + 2.11)


| Location | 13,587,617 – 13,587,714 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 70.91 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -13.84 |
| Energy contribution | -15.82 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
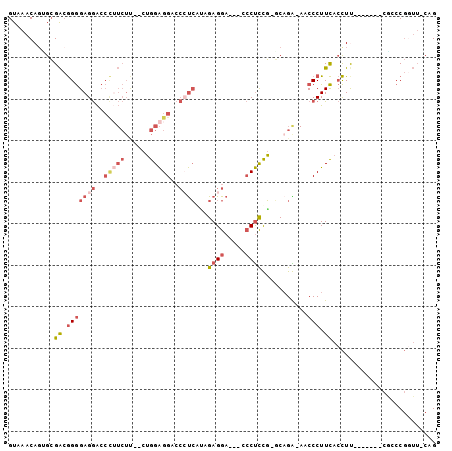
>3R_DroMel_CAF1 13587617 97 - 27905053 GUAAACAUAGCGACGGGGAGGACCCUUCUU--CUGGAGGACCCUCAAAGAGGA---CCCUCUG-GCAGAAAACCCUUCACCUUCACCCAUCGCCCGGUUCCAG ...(((...((((.(((((((....((((.--(..((((..((((...)))).---.))))..-).))))...))))........))).))))...))).... ( -31.40) >DroSec_CAF1 37110 91 - 1 GUAAACAGUGCGACGGGGAGGACCCAUCUU--CUGGAGGACCCUCAUGGAGGA---CCCUCUGGGCUGAAAACCCUUCACCUU-------CGCCCGGUUCCAG ...(((.(.((((.(((((((.....((.(--(..((((..((((...)))).---.))))..))..))....))))).)).)-------))).).))).... ( -31.60) >DroSim_CAF1 29251 91 - 1 GUAAACAGCGCGACGGGGAGGACCCUUCUU--CUGGAGGACCCUCACAGAGGA---CCCUCUGGGCAGAAAACCCUUCACCUU-------CGCCCGGUUCCAG ...(((.(.((((.(((((((....(((((--(..((((..((((...)))).---.))))..)).))))...))))).)).)-------))).).))).... ( -34.40) >DroEre_CAF1 27764 93 - 1 GUAAACAGUGCGACGGGGAGGACCCUUCUCUUCUGGAGGACCCUCGUUGAGGAGGGUCCUCCG-GCAGG-AACCCUUCACCUC-------CAGCCGCUU-CAG .........(((.(..(((((.......(((((((((((((((((......))))))))))))-).)))-)........))))-------).).)))..-... ( -43.36) >DroYak_CAF1 28086 88 - 1 GUAAACAGUGCGACGGGGAGGACCUUUCUC--CUGGAGGACCAUCAUUGAGGA---CCCUCCG-UCAGG-AACCCUUCACCUU-------CAACCGGUU-CAG ...........(((((((.((.((((....--.(((....))).....)))).---)))))))-))..(-((((.........-------.....))))-).. ( -26.84) >DroPer_CAF1 41109 73 - 1 GUAAACAGAGAGGGAGAGAGAG-----------AGGGGGGAGACGAGAGAGGA---GGCCUCU-UCAGG--ACCCUUUAGUUC-------CUGGC------AG .......(...(((((....((-----------((((.(....)((.(((((.---..)))))-))...--.))))))..)))-------))..)------.. ( -22.30) >consensus GUAAACAGUGCGACGGGGAGGACCCUUCUU__CUGGAGGACCCUCAUAGAGGA___CCCUCCG_GCAGA_AACCCUUCACCUU_______CGCCCGGUU_CAG ...........((.(((((((..(((((......)))))..))))...((((.....))))...........))).))......................... (-13.84 = -15.82 + 1.98)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:35 2006