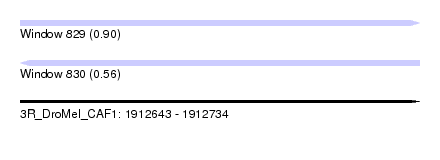
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,912,643 – 1,912,734 |
| Length | 91 |
| Max. P | 0.896984 |

| Location | 1,912,643 – 1,912,734 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 87.98 |
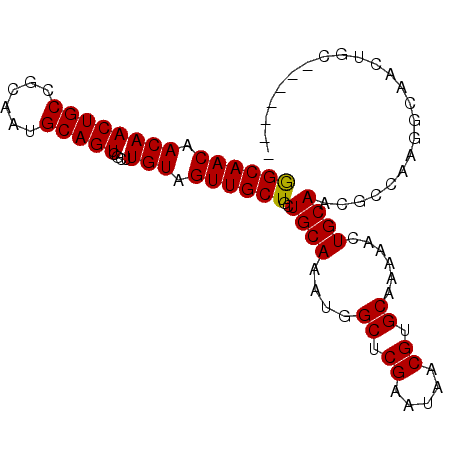
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -26.35 |
| Energy contribution | -26.93 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896984 |
| Prediction | RNA |
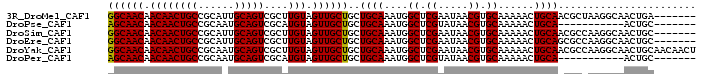
Download alignment: ClustalW | MAF
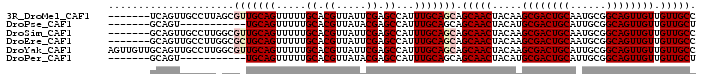
>3R_DroMel_CAF1 1912643 91 + 27905053 -------UCAGUUGCCUUAGCGUUGCAGUUUUUGCACGUUAUUCGAGCCAUUUGCAGCAGCAACUACAAGCGACUGCAAUGCGGCAGUUGUUGUUGCC -------.((..((((...(((((((((((..(((.((.....)).((.....)).)))((........)))))))))))))))))..))........ ( -29.00) >DroPse_CAF1 57443 80 + 1 -------GCAGU-----------UGCAGUUUUUGCACGUUAUACGAGCCAUUUGCAGCAGCAACUACAUGCGACUGCAUUGCGGCAGUUGUUGUUGCU -------...((-----------(((((.....((.((.....)).))...)))))))((((((.....((((((((......)))))))).)))))) ( -29.80) >DroSim_CAF1 71966 91 + 1 -------GCAGUUGCCUUGGCGUUGCAGUUUUUGCACGUUAUUCGAGCCAUUUGCAGCAGCAACUACAAGCGACUGCAAUGCGGCAGUUGUUGUUGCC -------...(((((..((((..(((((...)))))((.....)).))))...))))).(((((....(((((((((......)))))))))))))). ( -33.50) >DroEre_CAF1 73138 91 + 1 -------GCAGUUGCCUUGGCGCUGCAGUUUUUGCACGUUAUUCGAGCCAUUUGCAGCAGCAACUACAAGCGACUGCAAUGCGGCAGUUGUUGUUGCC -------((((((((.((((((((((((.....((.((.....)).))...))))))).)).....)))))))))))...(((((((...))))))). ( -33.70) >DroYak_CAF1 77024 98 + 1 AGUUGUUGCAGUUGCCUUGGCGUUGCAGUUUUUGCACGUUAUUCGAGCCAUUUGCAGCAGCAACUACAAGCGACUGCAUUGCGGCAGUUGUUGUUGCC .((.(((((.(((((..((((..(((((...)))))((.....)).))))...))))).))))).)).(((((((((......)))))))))...... ( -37.10) >DroPer_CAF1 57958 80 + 1 -------GCAGU-----------UGCAGUUUUUGCACGUUAUACGAGCCAUUUGCAGCAGCAACUACAUGCGACUGCAUUGCGGCAGUUGUUGUUGCU -------...((-----------(((((.....((.((.....)).))...)))))))((((((.....((((((((......)))))))).)))))) ( -29.80) >consensus _______GCAGUUGCCUUGGCGUUGCAGUUUUUGCACGUUAUUCGAGCCAUUUGCAGCAGCAACUACAAGCGACUGCAAUGCGGCAGUUGUUGUUGCC .....................(((((((.....((.((.....)).))...))))))).(((((.....((((((((......)))))))).))))). (-26.35 = -26.93 + 0.58)
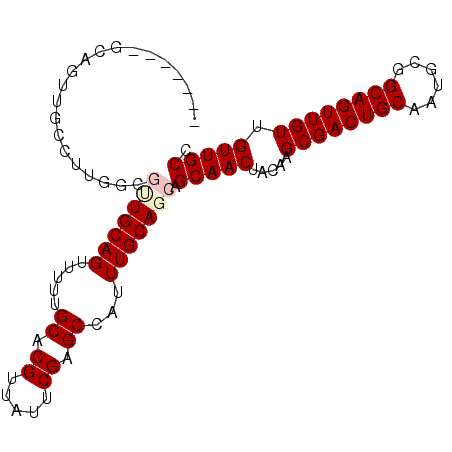
| Location | 1,912,643 – 1,912,734 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.98 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1912643 91 - 27905053 GGCAACAACAACUGCCGCAUUGCAGUCGCUUGUAGUUGCUGCUGCAAAUGGCUCGAAUAACGUGCAAAAACUGCAACGCUAAGGCAACUGA------- ((((........))))((.(((((((((.(((((((....))))))).))((.((.....)).))....))))))).))..((....))..------- ( -29.90) >DroPse_CAF1 57443 80 - 1 AGCAACAACAACUGCCGCAAUGCAGUCGCAUGUAGUUGCUGCUGCAAAUGGCUCGUAUAACGUGCAAAAACUGCA-----------ACUGC------- .(((.........((((...((((((.(((......))).))))))..))))..........((((.....))))-----------..)))------- ( -22.40) >DroSim_CAF1 71966 91 - 1 GGCAACAACAACUGCCGCAUUGCAGUCGCUUGUAGUUGCUGCUGCAAAUGGCUCGAAUAACGUGCAAAAACUGCAACGCCAAGGCAACUGC------- ((((........))))((.(((((((((.(((((((....))))))).))((.((.....)).))....))))))).))..((....))..------- ( -29.50) >DroEre_CAF1 73138 91 - 1 GGCAACAACAACUGCCGCAUUGCAGUCGCUUGUAGUUGCUGCUGCAAAUGGCUCGAAUAACGUGCAAAAACUGCAGCGCCAAGGCAACUGC------- ((((((.((((((((......)))))....))).))))))((((((....((.((.....)).))......))))))((....))......------- ( -30.70) >DroYak_CAF1 77024 98 - 1 GGCAACAACAACUGCCGCAAUGCAGUCGCUUGUAGUUGCUGCUGCAAAUGGCUCGAAUAACGUGCAAAAACUGCAACGCCAAGGCAACUGCAACAACU ((((........))))....((((((.(((((((((....)))))...((((.((.....))((((.....))))..)))))))).))))))...... ( -31.70) >DroPer_CAF1 57958 80 - 1 AGCAACAACAACUGCCGCAAUGCAGUCGCAUGUAGUUGCUGCUGCAAAUGGCUCGUAUAACGUGCAAAAACUGCA-----------ACUGC------- .(((.........((((...((((((.(((......))).))))))..))))..........((((.....))))-----------..)))------- ( -22.40) >consensus GGCAACAACAACUGCCGCAAUGCAGUCGCUUGUAGUUGCUGCUGCAAAUGGCUCGAAUAACGUGCAAAAACUGCAACGCCAAGGCAACUGC_______ ((((((.((((((((......)))))....))).))))))..((((....((.((.....)).))......))))....................... (-21.42 = -21.20 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:05 2006