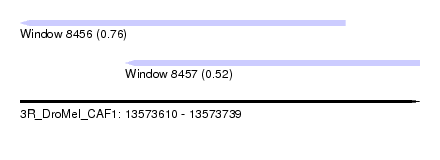
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,573,610 – 13,573,739 |
| Length | 129 |
| Max. P | 0.758120 |

| Location | 13,573,610 – 13,573,715 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.23 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13573610 105 - 27905053 AAAAAUGAAGUCAAGGCUGAGCGGGGAAAUGCGCCAUGCAAUGGGCUCUACUCCCCC-CAGCAUUUCCCCCCC----CCCUCUCUUUACCAAUAAAUUAAGACUCCUCGA .....(((((...(((..(.(.((((((((((((...))..((((..........))-)))))))))))))).----.)))..)))))...................... ( -27.70) >DroSim_CAF1 14615 99 - 1 AAAAAUGGAGUCAAGGCUGCGCGGGGAAAUGCGCCAUGCAAUGGGCUCUACUCCCCCGCAGCAUUUUUCGCC---------CUCUUUUCCAAUAAAUUAAGACUUCUC-- ......((((((...((((((.(((((...(.(((........))).)...)))))))))))..........---------...(((......)))....))))))..-- ( -32.80) >DroEre_CAF1 14146 94 - 1 AAAAAUGAAGCCAAGGCUGCGCUGGAAAGUGCGCUAUGCAAUGGGGUCUACUCCCC--CGGCGUUUCCC-----------CCGCAUUUCCAAUAAAUUAAAACUUCC--- ......((((....(((.(((((....))))))))((((...((((...((.((..--.)).))..)))-----------).))))................)))).--- ( -25.60) >DroYak_CAF1 14127 108 - 1 AAAAAUUAAGUCAAGGCUGCGCUGGGAAUUGCGCUAUGCAAUGGGGUCUACUCCCA--CAGCGUUUCCCUCGCAUUUCACCCGCAUUUCCAAUAAAUUAAAACUUCUCUG ..............((.((((..(((((.((((..((((..(((((.....)))))--..))))......)))).))).))))))...)).................... ( -24.70) >consensus AAAAAUGAAGUCAAGGCUGCGCGGGGAAAUGCGCCAUGCAAUGGGCUCUACUCCCC__CAGCAUUUCCC_CC________CCGCAUUUCCAAUAAAUUAAAACUUCUC__ ........(((....))).....(((((((((((...))...(((.......))).....)))))))))......................................... (-15.60 = -15.23 + -0.38)

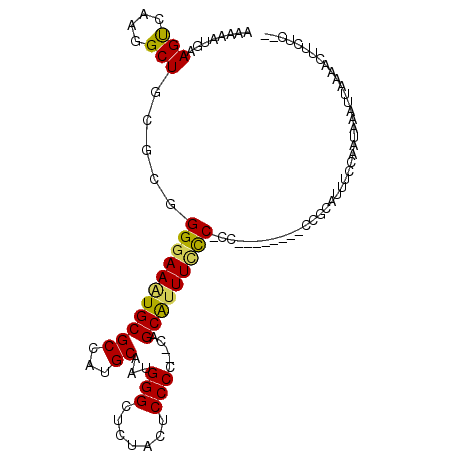

| Location | 13,573,644 – 13,573,739 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -17.35 |
| Energy contribution | -16.98 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13573644 95 - 27905053 UACCACUUGCAAUGAUAAAUCACAAAAAAUGAAGUCAAGGCUGAGCGGGGAAAUGCGCCAUGCAAUGGGCUCUACUCCCCC-CAGCAUUUCCCCCC .....(((((..((((...(((.......))).))))..)).))).((((((((((((...))..((((..........))-)))))))))))).. ( -27.10) >DroSim_CAF1 14642 96 - 1 UACCACUUGCAAUGAUAAAUCACAAAAAAUGGAGUCAAGGCUGCGCGGGGAAAUGCGCCAUGCAAUGGGCUCUACUCCCCCGCAGCAUUUUUCGCC ..(((.((....(((....)))....)).))).......((((((.(((((...(.(((........))).)...))))))))))).......... ( -30.20) >DroEre_CAF1 14173 91 - 1 UACCACUCGCACUGAUAAAUCACAAAAAAUGAAGCCAAGGCUGCGCUGGAAAGUGCGCUAUGCAAUGGGGUCUACUCCCC--CGGCGUUUCCC--- ..............................(((((...(((.(((((....))))))))..((...((((......))))--..)))))))..--- ( -24.90) >DroYak_CAF1 14165 94 - 1 UACCACUUGCAAUGAUAAAUCACAAAAAAUUAAGUCAAGGCUGCGCUGGGAAUUGCGCUAUGCAAUGGGGUCUACUCCCA--CAGCGUUUCCCUCG ......................................((..((((((...(((((.....)))))((((.....)))).--))))))..)).... ( -20.90) >consensus UACCACUUGCAAUGAUAAAUCACAAAAAAUGAAGUCAAGGCUGCGCGGGGAAAUGCGCCAUGCAAUGGGCUCUACUCCCC__CAGCAUUUCCC_CC ........((..(((....)))..........(((....)))..)).(((((((((((...))...(((.......))).....)))))))))... (-17.35 = -16.98 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:25 2006