
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,563,602 – 13,563,737 |
| Length | 135 |
| Max. P | 0.984203 |

| Location | 13,563,602 – 13,563,706 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.15 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -18.77 |
| Energy contribution | -18.27 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
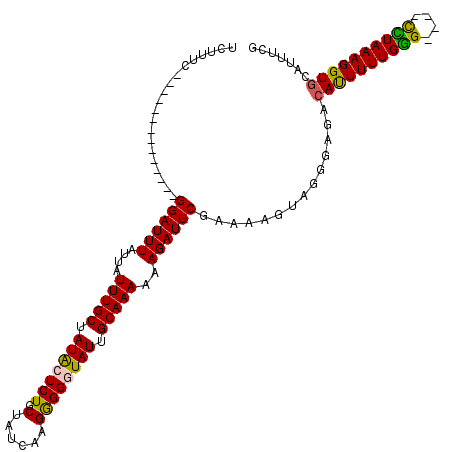
>3R_DroMel_CAF1 13563602 104 - 27905053 UCUUUC--------------GGAUUUAUUAUUUGCUAUACUCUGCUAUCAAGGGGGUAUUGCAAAAUAGAUUCGAAAAGUAGUGAGACAUUUUUGAGUGUGUUUAAAGGUGUAUUUCG ..((((--------------(((((((((..((((.(((((((.((....))))))))).)))))))))))))))))......((.(((((((((((....)))))))))))...)). ( -30.00) >DroSec_CAF1 5749 100 - 1 UAUUUC--------------GGAUUUAUUAUUUGCUAUACUCUGCUAUCAAGGGGUUAUUGCAAAAAAGAUUCGAAAAGUUGGGAGACAUUUUUGGG----UCUAAAGGUGCAUUUCG ..((((--------------((((((....(((((...(((((........)))))....)))))..))))))))))....(..(..((((((((..----..))))))))..)..). ( -24.50) >DroSim_CAF1 3707 100 - 1 UGUUUC--------------GGAUUUGUUAUUUGCUAUACUCUGCUAUCAAGGGGUUAUGGCAAAAAAGAUUCCAAAAGUAGGGAGACAUUUUUGGG----CCUAAAGGUGCAUUUCG ......--------------.((..(((..(((((((((((((........)))).))))))))).......((((((((..(....)))))))))(----((....))))))..)). ( -26.10) >DroEre_CAF1 4603 105 - 1 UCUUUCGCCUCACACCGUACGGAUUUAUUAUUUGCUAUGCUCUGCUUUUGUGAGGGUAUUGCAAA-GAGAUUCGAAAAGGUGCUCGACACUUUUGGG----CUUAAAGGU-------- (((((.((((.........(((((((....(((((.(((((((((....)).))))))).)))))-.))))))).(((((((.....))))))))))----)..))))).-------- ( -31.20) >consensus UCUUUC______________GGAUUUAUUAUUUGCUAUACUCUGCUAUCAAGGGGGUAUUGCAAAAAAGAUUCGAAAAGUAGGGAGACAUUUUUGGG____CCUAAAGGUGCAUUUCG ....................((((((....(((((.(((((((.(......)))))))).)))))..))))))..............((((((((((....))))))))))....... (-18.77 = -18.27 + -0.50)

| Location | 13,563,642 – 13,563,737 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -12.20 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13563642 95 - 27905053 AAAAUUCUUUCAAUGCCUUACAUUUUUUAUUUCUUUC--------------GGAUUUAUUAUUUGCUAUACUCUGCUAUCAAGGGGGUAUUGCAAAAUAGAUUCGAAAA .................................((((--------------(((((((((..((((.(((((((.((....))))))))).))))))))))))))))). ( -22.60) >DroSec_CAF1 5785 95 - 1 AAAAUUCUUCCAAUGCCUUAUAUUUUUUAUUUAUUUC--------------GGAUUUAUUAUUUGCUAUACUCUGCUAUCAAGGGGUUAUUGCAAAAAAGAUUCGAAAA .................................((((--------------((((((....(((((...(((((........)))))....)))))..)))))))))). ( -17.00) >DroSim_CAF1 3743 95 - 1 AAAAUUCUUCCAAUGCCUUACAUUUUUUAUUUGUUUC--------------GGAUUUGUUAUUUGCUAUACUCUGCUAUCAAGGGGUUAUGGCAAAAAAGAUUCCAAAA .....................................--------------(((.((....(((((((((((((........)))).)))))))))...)).))).... ( -16.90) >DroEre_CAF1 4631 108 - 1 AAAAUUCUUCCAAUGCCUUACAUUUUUUAUUUCUUUCGCCUCACACCGUACGGAUUUAUUAUUUGCUAUGCUCUGCUUUUGUGAGGGUAUUGCAAA-GAGAUUCGAAAA .................................(((((.(((...((....))........(((((.(((((((((....)).))))))).)))))-)))...))))). ( -20.60) >consensus AAAAUUCUUCCAAUGCCUUACAUUUUUUAUUUCUUUC______________GGAUUUAUUAUUUGCUAUACUCUGCUAUCAAGGGGGUAUUGCAAAAAAGAUUCGAAAA ...................................................((((((....(((((.(((((((.(......)))))))).)))))..))))))..... (-12.20 = -12.32 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:11 2006