
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,545,054 – 13,545,360 |
| Length | 306 |
| Max. P | 0.994210 |

| Location | 13,545,054 – 13,545,172 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.73 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13545054 118 - 27905053 UAUCACAGCAAGUCGAAAGCAGUGUACUGUAAUUCUC-GUAC-GUGUGCUGUUAAUGUCAGUAACAUUUUACUGUUAAGCGCAACUUCUCUUAUUAGCAAAUUGUGCAAGCAGUUCAAAA ....((((((.(((((..((((....)))).....))-).))-...)))))).(((((.....)))))..((((((..((((((.((((......)).)).)))))).))))))...... ( -25.70) >DroSec_CAF1 19829 119 - 1 UAGCACACCUAGUUGAAAGCAGUGUACUGUAAAUAUC-GUACAGUGUGCUGUAAAUAUCAGUAACAUUUUACUGUUAAGCGCAACUUCUCUUAUUAGUAAAUUGUGCAAGCAGUGUACUA (((.((((..........((((..(((((((......-.)))))))..))))........(((.((.(((((((.((((.(......).)))).))))))).))))).....)))).))) ( -31.50) >DroSim_CAF1 16851 119 - 1 UAUCACAGCUAGUCGAAAGCAGUGUACUGUAAAUAUC-GUACAGUGUGCUGUAAAUAUCAGUAACAUUUUACUGUUAAGCGCAACUUCUCUUAUUAGUAAAUUGUGCAAGCAGUGCACUA .......(((.......)))((((((((((.......-(((.(((((((((.......)))).))))).)))......((((((.((..((....))..))))))))..)))))))))). ( -33.30) >DroYak_CAF1 24727 99 - 1 UAUCAAAAC-------AAGCAGUGUACUGUAAGUACCAGUACAUUGUGCUGUAAAUGUUACUAACAUUUUACUGUUAAACGCAACUUCU--------------GCGCAAGCAGUGCACUA .......((-------(.(((((((((((.......)))))))))))..)))....(((..(((((......))))))))(((....((--------------((....))))))).... ( -29.40) >consensus UAUCACAGCUAGUCGAAAGCAGUGUACUGUAAAUAUC_GUACAGUGUGCUGUAAAUAUCAGUAACAUUUUACUGUUAAGCGCAACUUCUCUUAUUAGUAAAUUGUGCAAGCAGUGCACUA ..................((((..(((((((........)))))))..)))).................(((((((..((((((.................)))))).)))))))..... (-21.42 = -22.73 + 1.31)



| Location | 13,545,133 – 13,545,252 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.29 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13545133 119 + 27905053 AC-GAGAAUUACAGUACACUGCUUUCGACUUGCUGUGAUAUCGAGCAUUCCUGGUAUCGAGUGCAAAAUGGCGGCAUUUUAAGUUCAAUUUAACACGAUUAGGUUGAGCUUGUCGAUUCU ..-.((((...(((....)))...(((((.((((((.((.....((((((........))))))...)).))))))....(((((((((((((.....)))))))))))))))))))))) ( -34.40) >DroSec_CAF1 19909 119 + 1 AC-GAUAUUUACAGUACACUGCUUUCAACUAGGUGUGCUAUCGAGCAUUCAUGGUAUCGAAUGCAAAAUGGCGGCAUUUUAAGUUCAAUUUAUCACGAUUAGGUUGAGCUUGUCGGUUCU .(-(((......((((((((...........)))))))))))).((((((........)))))).......(((((.....(((((((((((.......))))))))))))))))..... ( -32.80) >DroSim_CAF1 16931 119 + 1 AC-GAUAUUUACAGUACACUGCUUUCGACUAGCUGUGAUAUCGAGCAUUCCUGGUAUCGAGUGCAAAAUGGCGGCAUUUUAAGUUCAAUUUAACACGAUUAGGUUGAGCUUGUCGGUUCU .(-((((((.(((((................)))))))))))).((((((........)))))).......(((((.....((((((((((((.....)))))))))))))))))..... ( -32.69) >DroYak_CAF1 24793 113 + 1 ACUGGUACUUACAGUACACUGCUU-------GUUUUGAUAUCGAGUAUUCCUGGUAUCGAGCAGAAAAUGGCGGCAUUUCUAGAUCAAUUUAACACGAUUAGAUUGAGCUUGCCAGUUUU (((((((...........(((((.-------.(((((...((((.(((.....))))))).)))))...))))).......((.(((((((((.....))))))))).)))))))))... ( -28.20) >consensus AC_GAUAUUUACAGUACACUGCUUUCGACUAGCUGUGAUAUCGAGCAUUCCUGGUAUCGAGUGCAAAAUGGCGGCAUUUUAAGUUCAAUUUAACACGAUUAGGUUGAGCUUGUCGGUUCU ...((.............(((((........(((.((((((((........))))))))))).......))))).....((((((((((((((.....))))))))))))))))...... (-22.10 = -22.29 + 0.19)

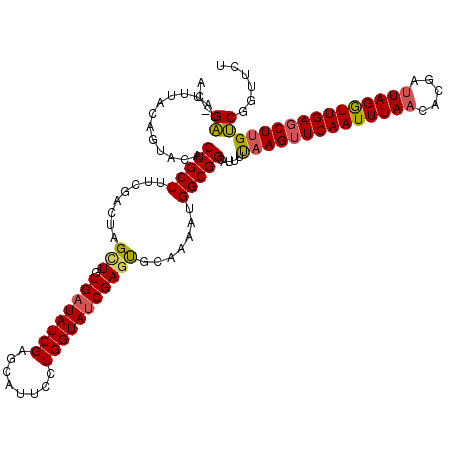
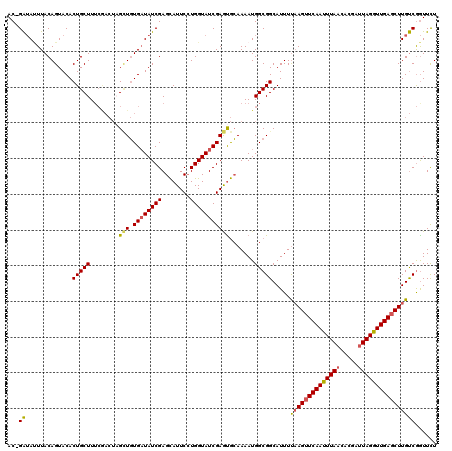
| Location | 13,545,252 – 13,545,360 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -18.14 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13545252 108 - 27905053 GCUGCCGCCUGCAACUCGCUCAGUUGCGUCACAAUAAGUAGCAACAAAAUAAAUAAAAUGCGACUCAUUAUGCGUUUAAUGAAACAUUUGUUGGCUUAUACUUAAAAA ..........((((((.....))))))......((((((..(((((((....((.(((((((........))))))).))......)))))))))))))......... ( -20.30) >DroSec_CAF1 20028 108 - 1 GCUGCAGCCUGCAACUCGCCCAGUUGCGUUACAAUAAGUAGCAACGAAAUAAAUAAAAUGCGACUCAUUUUGCGUUUAAGGAAACAUUUGUUGGCUUAUACUUAAAAA .....((((.((((((.....))))))(((((.....)))))((((((.......((((((((......))))))))..(....).))))))))))............ ( -27.20) >DroSim_CAF1 17050 108 - 1 GCUGCAGCCUGCAACUCGCCCAGUUGCGUCACAAUAAGUAGCAACGAAAUAAAUAAAAUGCGACUCAUUUUGCGUUUAAUGAAACAUUUGUUGGCUUAUACUUAUAAA (.((((((..((.....))...)))))).)...(((((((.(((((((....((.((((((((......)))))))).))......))))))).....)))))))... ( -23.30) >DroYak_CAF1 24906 108 - 1 GCUGCCGCCUGCAACUCGCCGAGUUGUGUCACAAGUAGCAGCAACAAAAUAAAUAAAGUGCGACACAUUAUGCCUUUAAUGAAACAUUUGUUGGCGUAUACUAAAAAU (((((..(.((((((((...)))))).....)).)..)))))...............(((...)))..((((((..(((........)))..)))))).......... ( -24.50) >consensus GCUGCAGCCUGCAACUCGCCCAGUUGCGUCACAAUAAGUAGCAACAAAAUAAAUAAAAUGCGACUCAUUAUGCGUUUAAUGAAACAUUUGUUGGCUUAUACUUAAAAA ((....))..((((((.....))))))......((((((..(((((((....((.((((((((......)))))))).))......)))))))))))))......... (-18.14 = -19.08 + 0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:03 2006