
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,542,128 – 13,542,428 |
| Length | 300 |
| Max. P | 0.999953 |

| Location | 13,542,128 – 13,542,227 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -10.79 |
| Energy contribution | -12.71 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13542128 99 - 27905053 GCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGCAGCAACAGCAGCAUCCGCCACCACCCCAUAUGCAGCAACAGC------AGCACCACGGCGGACCUGCACCA (((((.....((---((...........))))...)))))....((((..((((((...........(((.((....))------.)))....)))))).)))).... ( -28.26) >DroVir_CAF1 6802 102 - 1 GCAACAGCUAUGUCACACUAUCACCAUUUACAUCAACAACAACAG---CAGCCACAGGGGCCGCCUCUUCCACCACACA---UGCAGCACCACGGUGGACCACCGCCA ((((((....)))...............................(---(.(((.....))).))...............---)))........(((((....))))). ( -17.40) >DroSec_CAF1 16874 99 - 1 GCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGCAACAACAGCAGCAUCCGCCACCACCCCAUAUGCAGCAACAGC------AGCACCACGGCGGACCUGCACCA (((((((..(((---......)))...))))...))).......((((..((((((...........(((.((....))------.)))....)))))).)))).... ( -25.46) >DroSim_CAF1 13888 99 - 1 GCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGCAGCAACAGCAGCAUCCGCCACCACCCCAUAUGCAGCAACAGC------AGCACCACGGCGGACCUGCACCA (((((.....((---((...........))))...)))))....((((..((((((...........(((.((....))------.)))....)))))).)))).... ( -28.26) >DroYak_CAF1 20680 93 - 1 GCUGCAGUCAUG---CACUAUCAUCAUUUACACCAG---------CAGCAUCCGCCACCACCCCAUAUGCAGCAACAGC---AGCAGCACCACGGCGGACCUGCACCA ..((((....))---))..................(---------(((..((((((...........(((.((....))---.)))(.....))))))).)))).... ( -23.60) >DroAna_CAF1 14590 102 - 1 GCUGCUGUCAUG---CAUUAUCAUCACCUGCACCAGCAGC---AGCAGCACCCGCCGCCGCCUCACAUGCAGCAACAACAACAGCCGCACCACGGCGGACCGGCGCCA ((((((((..((---((...........))))...)))))---)))......((((((((((.....(((.((..........)).)))....)))))..)))))... ( -38.70) >consensus GCUGCAGUCAUG___CACUAUCAUCAUCUGCACCAGCAGCAACAGCAGCAUCCGCCACCACCCCAUAUGCAGCAACAGC______AGCACCACGGCGGACCUGCACCA ((((((((........)))........((((....)))).....))))).((((((.....................................))))))......... (-10.79 = -12.71 + 1.92)

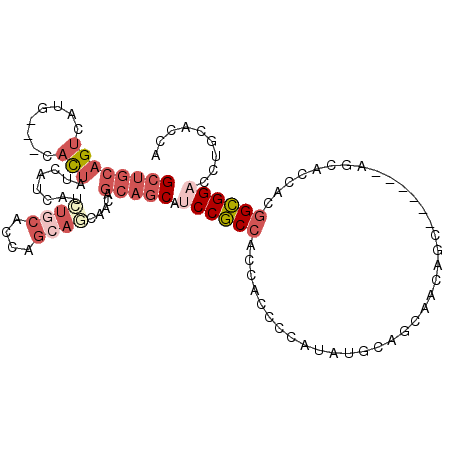

| Location | 13,542,162 – 13,542,267 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.09 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -21.83 |
| Energy contribution | -24.88 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13542162 105 + 27905053 AUAUGGGGUGGUGGCGGAUGCUGCUGUUGCUGCUGGUGCAGAUGAUGAUAGUG---CAUGACUGCAGCUGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGG ......((..(..((((......))))..)..))(.(((((.(.(((......---))).)))))).).(((((.((((((((.......)))))))).))))).... ( -46.40) >DroVir_CAF1 6839 87 + 1 AGAGGCGGCCCCUGUGGCUG---CUGUUGUUGUUGAUGUAAAUGGUGAUAGUGUGACAUAGCUGUUGCUGAGUCGG------GAGUUGA------------ACCUGGC ((..(((((...(((.((.(---((((..(..((......))..)..))))))).)))..)))))..))..(((((------(......------------.)))))) ( -28.90) >DroSec_CAF1 16908 105 + 1 AUAUGGGGUGGUGGCGGAUGCUGCUGUUGUUGCUGGUGCAGAUGAUGAUAGUG---CAUGACUGCAGCUGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGG ......((..(..((((......))))..)..))(.(((((.(.(((......---))).)))))).).(((((.((((((((.......)))))))).))))).... ( -44.50) >DroSim_CAF1 13922 105 + 1 AUAUGGGGUGGUGGCGGAUGCUGCUGUUGCUGCUGGUGCAGAUGAUGAUAGUG---CAUGACUGCAGCUGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGG ......((..(..((((......))))..)..))(.(((((.(.(((......---))).)))))).).(((((.((((((((.......)))))))).))))).... ( -46.40) >DroYak_CAF1 20717 96 + 1 AUAUGGGGUGGUGGCGGAUGCUG---------CUGGUGUAAAUGAUGAUAGUG---CAUGACUGCAGCUGGAUCCGGUCCUGGUGGUCCUCCGGGACCAGAUCCUGGG .........(((.((((((((.(---------(((..((.....))..)))))---)))..)))).)))(((((.((((((((.......)))))))).))))).... ( -35.50) >DroAna_CAF1 14630 96 + 1 AUGUGAGGCGGCGGCGGGUGCUGCU---GCUGCUGGUGCAGGUGAUGAUAAUG---CAUGACAGCAGCCGGGUC------UGGCGGACCGCCGGGUGCGGAGCCAGGG ......(((((((((....))))))---))).(((((((..((.(((......---))).)).))).)))).((------((((...((((.....)))).)))))). ( -43.20) >consensus AUAUGGGGUGGUGGCGGAUGCUGCUGUUGCUGCUGGUGCAGAUGAUGAUAGUG___CAUGACUGCAGCUGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGG ......((((((..(((......)))..))))))(.(((((.(.(((.........))).)))))).).(((((.((((((((.......)))))))).))))).... (-21.83 = -24.88 + 3.06)



| Location | 13,542,162 – 13,542,267 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.09 |
| Mean single sequence MFE | -31.49 |
| Consensus MFE | -15.34 |
| Energy contribution | -18.93 |
| Covariance contribution | 3.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.49 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13542162 105 - 27905053 CCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCAGCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGCAGCAACAGCAGCAUCCGCCACCACCCCAUAU ....((.(((((((((((.......))))))))))).)).((((((((....---.)))........((((....)))).....)))))................... ( -37.40) >DroVir_CAF1 6839 87 - 1 GCCAGGU------------UCAACUC------CCGACUCAGCAACAGCUAUGUCACACUAUCACCAUUUACAUCAACAACAACAG---CAGCCACAGGGGCCGCCUCU ....(((------------.......------..(((..(((....)))..)))........)))...................(---(.(((.....))).)).... ( -14.43) >DroSec_CAF1 16908 105 - 1 CCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCAGCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGCAACAACAGCAGCAUCCGCCACCACCCCAUAU ....((.(((((((((((.......))))))))))).)).((((((((....---.))).........(((....)))......)))))................... ( -35.20) >DroSim_CAF1 13922 105 - 1 CCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCAGCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGCAGCAACAGCAGCAUCCGCCACCACCCCAUAU ....((.(((((((((((.......))))))))))).)).((((((((....---.)))........((((....)))).....)))))................... ( -37.40) >DroYak_CAF1 20717 96 - 1 CCCAGGAUCUGGUCCCGGAGGACCACCAGGACCGGAUCCAGCUGCAGUCAUG---CACUAUCAUCAUUUACACCAG---------CAGCAUCCGCCACCACCCCAUAU ....(((((((((((.((.......)).))))))))))).(((((.......---....................)---------))))................... ( -30.73) >DroAna_CAF1 14630 96 - 1 CCCUGGCUCCGCACCCGGCGGUCCGCCA------GACCCGGCUGCUGUCAUG---CAUUAUCAUCACCUGCACCAGCAGC---AGCAGCACCCGCCGCCGCCUCACAU ..(((((.((((.....))))...))))------)....(((((((((..((---((...........))))...)))))---)((.((....)).)).)))...... ( -33.80) >consensus CCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCAGCUGCAGUCAUG___CACUAUCAUCAUCUGCACCAGCAGCAACAGCAGCAUCCGCCACCACCCCAUAU ....((.((((((((.((.......)).)))))))).)).((((((((........)))........((((....)))).....)))))................... (-15.34 = -18.93 + 3.60)



| Location | 13,542,193 – 13,542,297 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -43.90 |
| Consensus MFE | -25.72 |
| Energy contribution | -27.00 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13542193 104 + 27905053 GCUGGUGCAGAUGAUGAUAGUG---CAUGACUGCAGC---UGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGGGGCGGUGGUGCGCCGUAUGGAUGUGGGGCA- ((((...((.....)).))))(---(((.(((((..(---((((((.((((((((.......)))))))).))))).))..))))).))))(((.(((....))).))).- ( -49.70) >DroGri_CAF1 11154 93 + 1 GUUGAUGUAGAUGGUGAUAGUGGGACAUGGCUGCGGCAGCCGGAUCAU------UUGGUGC------------ACCUGGCGCUGGUGGCCCGCCAUAGGGAUGGGGCAAGG ......((....((((..(((((....((((((...))))))..))))------).....)------------)))..)).....((.((((((....)).)))).))... ( -27.80) >DroSec_CAF1 16939 104 + 1 GCUGGUGCAGAUGAUGAUAGUG---CAUGACUGCAGC---UGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGGGGCGGUGGUGCGCCGUAUGGAUGUGGGGCA- ((((...((.....)).))))(---(((.(((((..(---((((((.((((((((.......)))))))).))))).))..))))).))))(((.(((....))).))).- ( -49.70) >DroSim_CAF1 13953 104 + 1 GCUGGUGCAGAUGAUGAUAGUG---CAUGACUGCAGC---UGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGGGGCGGUGGUGCGCCGUAUGGAUGUGGGGCA- ((((...((.....)).))))(---(((.(((((..(---((((((.((((((((.......)))))))).))))).))..))))).))))(((.(((....))).))).- ( -49.70) >DroYak_CAF1 20740 103 + 1 -CUGGUGUAAAUGAUGAUAGUG---CAUGACUGCAGC---UGGAUCCGGUCCUGGUGGUCCUCCGGGACCAGAUCCUGGGGGCAGUGGUGCGCCAUAUGGAUGUGGCGCA- -..................(((---(((.(((((..(---((((((.((((((((.......)))))))).))))).))..))))).))))((((((....)))))))).- ( -49.10) >DroAna_CAF1 14658 98 + 1 GCUGGUGCAGGUGAUGAUAAUG---CAUGACAGCAGC---CGGGUC------UGGCGGACCGCCGGGUGCGGAGCCAGGGGGUGGUGGGGCACCAUAGGGGUGAGGCGCU- (((((((((...........))---)))..)))).((---(.((((------.....)))).((.(((((...((((.....))))...)))))...)).....)))...- ( -37.40) >consensus GCUGGUGCAGAUGAUGAUAGUG___CAUGACUGCAGC___UGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGGGGCGGUGGUGCGCCAUAUGGAUGUGGCGCA_ ((((.(((((.(.(((.........))).))))))......(((((.((((((((.......)))))))).)))))......))))..(((.(((((....))))).))). (-25.72 = -27.00 + 1.28)



| Location | 13,542,193 – 13,542,297 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -16.16 |
| Energy contribution | -18.15 |
| Covariance contribution | 1.99 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13542193 104 - 27905053 -UGCCCCACAUCCAUACGGCGCACCACCGCCCCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCA---GCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGC -.((.............((((......))))....((.(((((((((((.......))))))))))).)).---))(((((..(((---......)))...)))))..... ( -36.20) >DroGri_CAF1 11154 93 - 1 CCUUGCCCCAUCCCUAUGGCGGGCCACCAGCGCCAGGU------------GCACCAA------AUGAUCCGGCUGCCGCAGCCAUGUCCCACUAUCACCAUCUACAUCAAC ................(((((.........)))))(((------------(......------..(((..(((((...)))))..))).......))))............ ( -19.76) >DroSec_CAF1 16939 104 - 1 -UGCCCCACAUCCAUACGGCGCACCACCGCCCCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCA---GCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGC -.((.............((((......))))....((.(((((((((((.......))))))))))).)).---))(((((..(((---......)))...)))))..... ( -36.20) >DroSim_CAF1 13953 104 - 1 -UGCCCCACAUCCAUACGGCGCACCACCGCCCCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCA---GCUGCAGUCAUG---CACUAUCAUCAUCUGCACCAGC -.((.............((((......))))....((.(((((((((((.......))))))))))).)).---))(((((..(((---......)))...)))))..... ( -36.20) >DroYak_CAF1 20740 103 - 1 -UGCGCCACAUCCAUAUGGCGCACCACUGCCCCCAGGAUCUGGUCCCGGAGGACCACCAGGACCGGAUCCA---GCUGCAGUCAUG---CACUAUCAUCAUUUACACCAG- -(((((((........)))))))..(((((.....(((((((((((.((.......)).))))))))))).---...)))))....---.....................- ( -40.00) >DroAna_CAF1 14658 98 - 1 -AGCGCCUCACCCCUAUGGUGCCCCACCACCCCCUGGCUCCGCACCCGGCGGUCCGCCA------GACCCG---GCUGCUGUCAUG---CAUUAUCAUCACCUGCACCAGC -...(((.........(((((...)))))....(((((.((((.....))))...))))------)....)---)).((((...((---((...........)))).)))) ( -30.10) >consensus _UGCCCCACAUCCAUACGGCGCACCACCGCCCCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCA___GCUGCAGUCAUG___CACUAUCAUCAUCUGCACCAGC .(((.............((((......))))....((.((((((((.((.......)).)))))))).)).......)))............................... (-16.16 = -18.15 + 1.99)

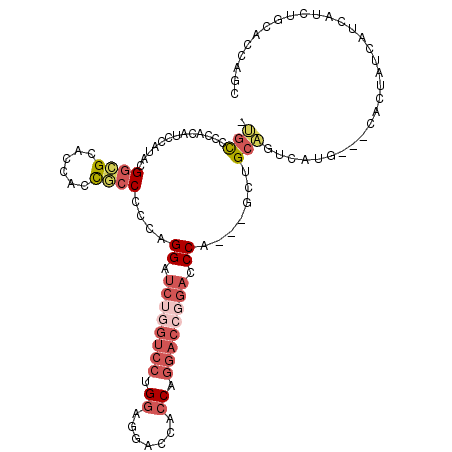

| Location | 13,542,227 – 13,542,323 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 75.69 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -31.55 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13542227 96 + 27905053 UGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGGGGCGGUGGUGCGCCGUAUGGAUGUGGGGCAACAUUUUGUUGUUGCUGCUGGAUGCU .(.((((((((((((.......))))).(((.((((..(.((((......)))).)..)))).)))(((((((......)))))))))))))).). ( -45.40) >DroSim_CAF1 13987 96 + 1 UGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGGGGCGGUGGUGCGCCGUAUGGAUGUGGGGCAACAUUUUGUUGUUGCUGCUGGAUGCU .(.((((((((((((.......))))).(((.((((..(.((((......)))).)..)))).)))(((((((......)))))))))))))).). ( -45.40) >DroAna_CAF1 14692 84 + 1 CGGGUC------UGGCGGACCGCCGGGUGCGGAGCCAGGGGGUGGUGGGGCACCAUAGGGGUGAGGCGCUAUACUCU------GCUGCUGGACGCU .(.(((------..((((.((((.....))))...(((((..(((((...((((.....))))...)))))..))))------)))))..))).). ( -40.00) >consensus UGGGUCCGGUCCUGGUGGUCCUCCAGGACCAGAUCCUGGGGGCGGUGGUGCGCCGUAUGGAUGUGGGGCAACAUUUUGUUGUUGCUGCUGGAUGCU .(.(((((((...(((.((((.((((((.....))))))))))((((.(((.(((((....))))).))).))))........)))))))))).). (-31.55 = -31.90 + 0.35)

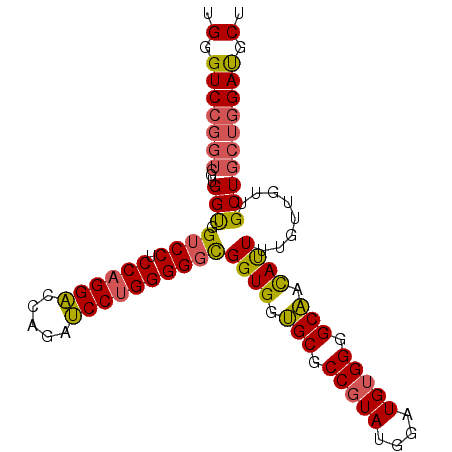

| Location | 13,542,227 – 13,542,323 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 75.69 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -25.44 |
| Energy contribution | -28.67 |
| Covariance contribution | 3.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.77 |
| SVM decision value | 4.82 |
| SVM RNA-class probability | 0.999953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13542227 96 - 27905053 AGCAUCCAGCAGCAACAACAAAAUGUUGCCCCACAUCCAUACGGCGCACCACCGCCCCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCA .((.....)).((((((......)))))).............((((......))))....((.(((((((((((.......))))))))))).)). ( -37.20) >DroSim_CAF1 13987 96 - 1 AGCAUCCAGCAGCAACAACAAAAUGUUGCCCCACAUCCAUACGGCGCACCACCGCCCCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCA .((.....)).((((((......)))))).............((((......))))....((.(((((((((((.......))))))))))).)). ( -37.20) >DroAna_CAF1 14692 84 - 1 AGCGUCCAGCAGC------AGAGUAUAGCGCCUCACCCCUAUGGUGCCCCACCACCCCCUGGCUCCGCACCCGGCGGUCCGCCA------GACCCG .((.....))...------........(((((..........)))))...........(((((.((((.....))))...))))------)..... ( -24.20) >consensus AGCAUCCAGCAGCAACAACAAAAUGUUGCCCCACAUCCAUACGGCGCACCACCGCCCCCAGGAUCUGGUCCUGGAGGACCACCAGGACCGGACCCA .((.....)).((((((......)))))).............((((......))))....((.(((((((((((.......))))))))))).)). (-25.44 = -28.67 + 3.22)

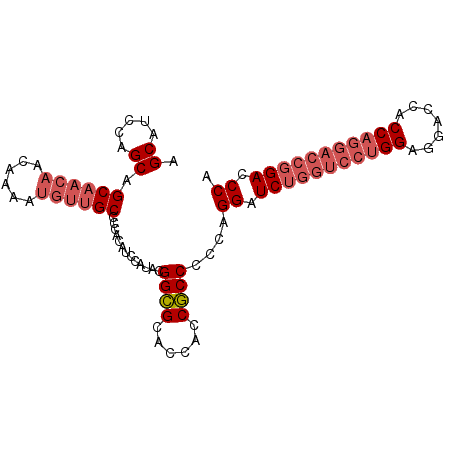
| Location | 13,542,323 – 13,542,428 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 68.52 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.68 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13542323 105 - 27905053 CCUGCUGC------CACGCCCCCAUCGGCUGGAGCGGCUCCUGGUGCCGCUACACCGCCAACCUCGGGCCCGCCCACACCCAACAACAACAGCAACAACGGUAGCGAUCCC ..((((((------(..(((......)))(((.(((((((..(((...((......))..)))..))).))))))).......................)))))))..... ( -34.90) >DroVir_CAF1 6985 99 - 1 GCGACUGGUGCAAGCGCAUCCCCAACUGUGGCAGCCAGUGCAGGUGGCGCUACACCACCAACCGCGGGUCCACCCACACCGAA------------CAACGGCAACGAUUCU (((..(((((..(((((..((...((((.(....)))))...))..)))))....)))))..)))(((....)))........------------...((....))..... ( -33.80) >DroSim_CAF1 14083 105 - 1 CCUGCUGC------CACGCCCCCAUCGGCUGGAGCGGCACCUGGUGGCGCUACACCGCCAACCUCGGGCCCGCCCACGCCCAACAAUAACAGCAACAACGGCAGCGAUCCC ..((((((------(..(((......)))(((.((((..((.(((((((......)))).)))..))..))))))).......................)))))))..... ( -40.90) >DroAna_CAF1 14776 93 - 1 CCCUCUGC------CACGCCCCCGUCGGC------GGCA------GGUGCCACACCGCCAACUUCGGGCCCGCCCACUCCCAAUAAUAACAGUAACAACGGCAGUGACCCC ..(.((((------(..(((......)))------(((.------((((...)))))))......(((....)))........................))))).)..... ( -28.40) >consensus CCUGCUGC______CACGCCCCCAUCGGCUGGAGCGGCACCUGGUGGCGCUACACCGCCAACCUCGGGCCCGCCCACACCCAACAAUAACAGCAACAACGGCAGCGAUCCC ................(((..((...(((.(((.....))).((((......)))))))......(((....)))........................))..)))..... (-15.93 = -16.68 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:57 2006