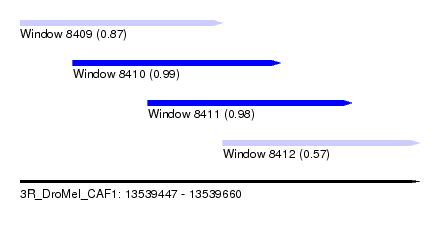
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,539,447 – 13,539,660 |
| Length | 213 |
| Max. P | 0.993903 |

| Location | 13,539,447 – 13,539,555 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -12.06 |
| Energy contribution | -13.06 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13539447 108 + 27905053 CUGC------------GGCGGGUUAGCCCCCUAACUACCACACUACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGAAAAAAUCAUAUCACGUGCUUCUCCAUU ..((------------((.(((....))))).............................((((((.(((...))).))))))....((((((..........))))))))......... ( -18.30) >DroSec_CAF1 14256 102 + 1 CUGC---------------GGGUAAGCCCCCUAACUACCACACUACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA---AAUCAUAUCACGUGCUUCUCCAUU ....---------------((..((((.................................((((((.(((...))).))))))....((((((---.......))))))))))..))... ( -17.90) >DroSim_CAF1 11209 102 + 1 CUGC---------------GGGUAAGCCCCCUAACUACCACACUUCCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA---AAUCAUAUCACGUGCUUCUCCAUU ....---------------((..((((.................................((((((.(((...))).))))))....((((((---.......))))))))))..))... ( -17.90) >DroYak_CAF1 18137 102 + 1 CUGC---------------GGGUAAGCCCCCGAACUAUAACUCUACCCCACUAAUGAAACAAUUAACCGGUUUAUAUUUAAUUCAAAACGUGA---AAUCACAUCACGUGCUUCUCCUUU ....---------------((..((((....(((.((.......(((....((((......))))...))).......)).)))...((((((---.......))))))))))..))... ( -16.94) >DroAna_CAF1 12189 114 + 1 CUGCUAAUAUAUGUAUGGUAUGUGGGCUCUCCACCUCGCUCACUGCCUCGCCAAUAAAACAAUUAACCGGUUUCCGUUUAAAUGGAAACGUGA---AGUCGCAUCACGUGCACC---GAG ............((..((((.((((((..........))))))))))..))................((((((((((....))))))((((((---.......))))))..)))---).. ( -28.80) >consensus CUGC_______________GGGUAAGCCCCCUAACUACCACACUACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA___AAUCAUAUCACGUGCUUCUCCAUU ...................(((.((((.................................((((((.((.....)).))))))....((((((..........)))))))))).)))... (-12.06 = -13.06 + 1.00)

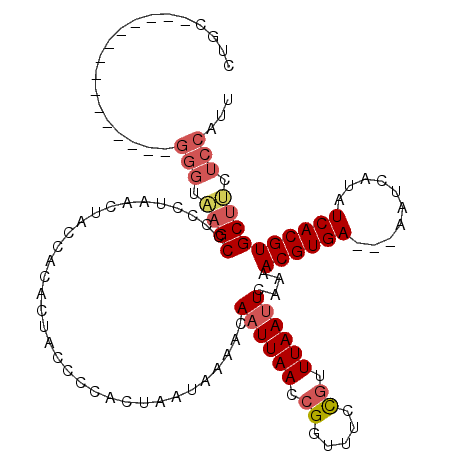
| Location | 13,539,475 – 13,539,586 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -19.46 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13539475 111 + 27905053 CACUACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGAAAAAAUCAUAUCACGUGCUUCUCCAUUGGGGCC---AACAAACCAAGCA------AACCAAACCAAA .....(((((..........((((((.(((...))).))))))....((((((..........))))))..........)))))..---.............------............ ( -18.00) >DroSec_CAF1 14281 108 + 1 CACUACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA---AAUCAUAUCACGUGCUUCUCCAUUGGGGCC---AAUAAACCAAGCA------AACCAAACCAAA .....(((((..........((((((.(((...))).))))))....((((((---.......))))))..........)))))..---.............------............ ( -18.60) >DroSim_CAF1 11234 108 + 1 CACUUCCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA---AAUCAUAUCACGUGCUUCUCCAUUGGGGCC---AAUAAACCAAGCA------AACCAAACCAAA .....(((((..........((((((.(((...))).))))))....((((((---.......))))))..........)))))..---.............------............ ( -18.20) >DroYak_CAF1 18162 114 + 1 CUCUACCCCACUAAUGAAACAAUUAACCGGUUUAUAUUUAAUUCAAAACGUGA---AAUCACAUCACGUGCUUCUCCUUUGGGGCC---AAUAAACCAGGCAAAAAAAAACCAAACCAAA .......((..((((......))))...(((((((............((((((---.......))))))(((((......))))).---.))))))).)).................... ( -19.70) >DroAna_CAF1 12229 95 + 1 CACUGCCUCGCCAAUAAAACAAUUAACCGGUUUCCGUUUAAAUGGAAACGUGA---AGUCGCAUCACGUGCACC---GAGCGGGCCGGCCAUCAACCAAAC------------------- ..(.(((.(((.(((......)))...((((((((((....))))))((((((---.......))))))..)))---).)))))).)..............------------------- ( -22.80) >consensus CACUACCCCACUAAUAAAACAAUUAACCGGUUUCCGUUUAAUUCAAAACGUGA___AAUCAUAUCACGUGCUUCUCCAUUGGGGCC___AAUAAACCAAGCA______AACCAAACCAAA .....(((((..........((((((.((.....)).))))))....((((((..........))))))..........))))).................................... (-14.10 = -14.58 + 0.48)

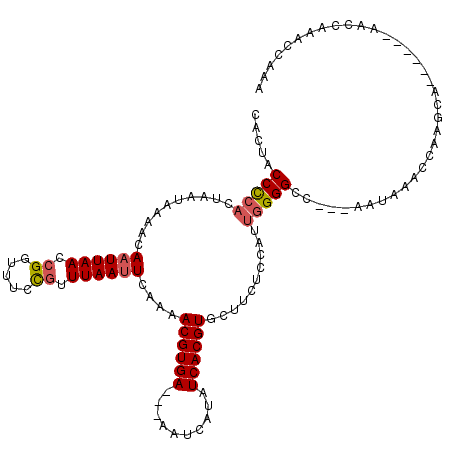
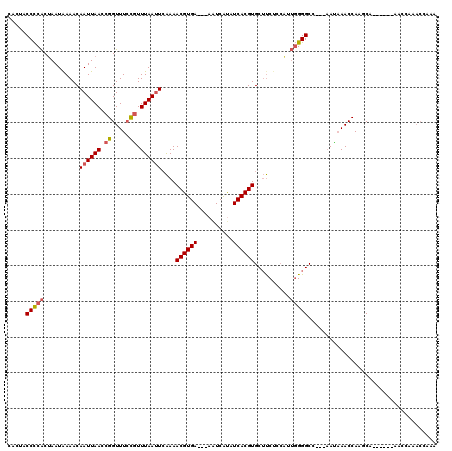
| Location | 13,539,515 – 13,539,624 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -16.23 |
| Energy contribution | -16.97 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13539515 109 + 27905053 AUUCAAAACGUGAAAAAAUCAUAUCACGUGCUUCUCCAUUGGGGCCAACAAACCAAGCA------AACCAAACCAAACACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAU ..(((..((((((..........))))))(((((......)))))..............------....................(((((((......)))))))......))). ( -19.30) >DroSec_CAF1 14321 106 + 1 AUUCAAAACGUGA---AAUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCA------AACCAAACCAAAAACCAACUUCAAGCGCUUUAUCGCUUGACACAACUGAU ..(((..((((((---.......))))))((((....((((....)))).....)))).------....................(((((((......)))))))......))). ( -20.20) >DroSim_CAF1 11274 106 + 1 AUUCAAAACGUGA---AAUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCA------AACCAAACCAAAAACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAU ..(((..((((((---.......))))))((((....((((....)))).....)))).------....................(((((((......)))))))......))). ( -20.20) >DroYak_CAF1 18202 108 + 1 AUUCAAAACGUGA---AAUCACAUCACGUGCUUCUCCUUUGGGGCCAAUAAACCAGGCAAAAAAAAACCAAACCAAAAACCAACUUCAA----CUUAUCGCUUGACACAACUGAU ..(((..((((((---.......))))))(((((......)))))........(((((...............................----......))))).......))). ( -14.49) >consensus AUUCAAAACGUGA___AAUCAUAUCACGUGCUUCUCCAUUGGGGCCAAUAAACCAAGCA______AACCAAACCAAAAACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAU ..(((..((((((..........))))))(((((......)))))........................................(((((((......)))))))......))). (-16.23 = -16.97 + 0.75)

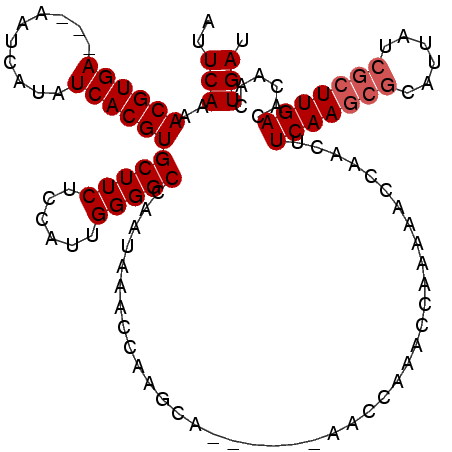
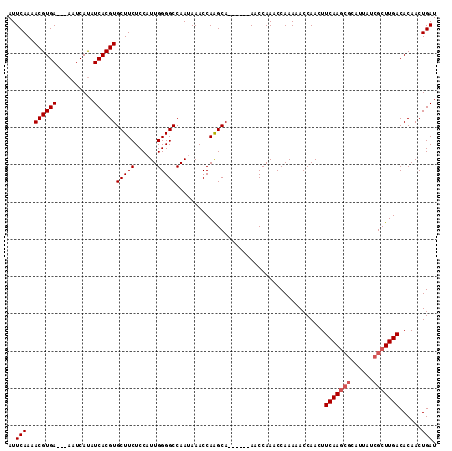
| Location | 13,539,555 – 13,539,660 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -13.10 |
| Energy contribution | -15.35 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13539555 105 + 27905053 GGGGCCAACAAACCAAGCA------AACCAAACCAAACACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAUAGGCAAAUUCUUGAUAUCAGGGAUUACAAAGAGUAA---- ((.((...........)).------..))..........((....(((((((......))))))).....((((((..(((....))).)))))))).((((.....))))---- ( -20.50) >DroSec_CAF1 14358 108 + 1 GGGGCCAAUAAACCAAGCA------AACCAAACCAAAAACCAACUUCAAGCGCUUUAUCGCUUGACACAACUGAUA-GCAAAUUCUUCAUAUCAGGGAUUACAAUGAGUAAUCCU ((.((...........)).------..))................(((((((......))))))).....((((((-............))))))(((((((.....))))))). ( -24.40) >DroSim_CAF1 11311 108 + 1 GGGGCCAAUAAACCAAGCA------AACCAAACCAAAAACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAUA-GCAAAUUCUUCAUAUCAGGGAUUACAAUGAGUAAUCCU ((.((...........)).------..))................(((((((......))))))).....((((((-............))))))(((((((.....))))))). ( -24.40) >DroYak_CAF1 18239 110 + 1 GGGGCCAAUAAACCAGGCAAAAAAAAACCAAACCAAAAACCAACUUCAA----CUUAUCGCUUGACACAACUGAUA-GCAAGUUCUUGAUAUCAGGGAUUACAAUAAAUAAGACU ...(((.........)))...............................----(((((...(((......((((((-.(((....))).))))))......)))...)))))... ( -15.00) >consensus GGGGCCAAUAAACCAAGCA______AACCAAACCAAAAACCAACUUCAAGCGCAUUAUCGCUUGACACAACUGAUA_GCAAAUUCUUCAUAUCAGGGAUUACAAUGAGUAAUCCU .((.........))...............................(((((((......))))))).....((((((.............))))))(((((((.....))))))). (-13.10 = -15.35 + 2.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:42 2006