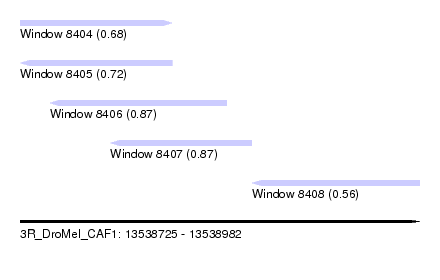
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,538,725 – 13,538,982 |
| Length | 257 |
| Max. P | 0.874618 |

| Location | 13,538,725 – 13,538,823 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -19.46 |
| Energy contribution | -19.27 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13538725 98 + 27905053 CCUCCACCACCGUC-CCUUCUUCUUCUGCUUCUCCUACCCCAAAACAGAGAGAGGGAGGGAGAAUCAGCUGUGACAACAACAAGUUUCUCUGUUAUUGC ............((-(((((((((((((.................)))).)))))))))))......((.((((((..............)))))).)) ( -23.47) >DroSec_CAF1 13583 98 + 1 CCUCCACCUCCGCC-CCUUCUUCUUCUGCUUCUCCUACCCCAAAACAGAGAGAGGGAGGGAGAAUCAGCUGUGACAACAACAAGUUUCUCUGUUAUUGC .............(-(((((((((((((.................)))).)))))))))).......((.((((((..............)))))).)) ( -21.97) >DroSim_CAF1 10510 99 + 1 CCUCCACCACCGCCCCCUUCUUCUUCUGCUUCUCCUACCCCAAAACAGAGAGAGGGAGGGAGAAUCAGCUGUGACAACAACAAGUUUCUCUGUUAUUGC .......(((.((....(((((((((..((((((((..........)).)))))))))))))))...)).))).(((.((((........)))).))). ( -23.10) >DroYak_CAF1 17408 95 + 1 CCUCCUUCUCCACC----UCUUUUUCUGUUUCUCCUACCCCAAAACAGAGAGAGGGAGGGAGAAUCAGCUGUGACAACAACAAGUUUCUCUGUUAUUGC .....((((((..(----(((((((((((((...........))))))))))))))..))))))...((.((((((..............)))))).)) ( -30.14) >consensus CCUCCACCACCGCC_CCUUCUUCUUCUGCUUCUCCUACCCCAAAACAGAGAGAGGGAGGGAGAAUCAGCUGUGACAACAACAAGUUUCUCUGUUAUUGC ..................((..(..(((.(((((((.((((..........).))).))))))).)))..).))(((.((((........)))).))). (-19.46 = -19.27 + -0.19)

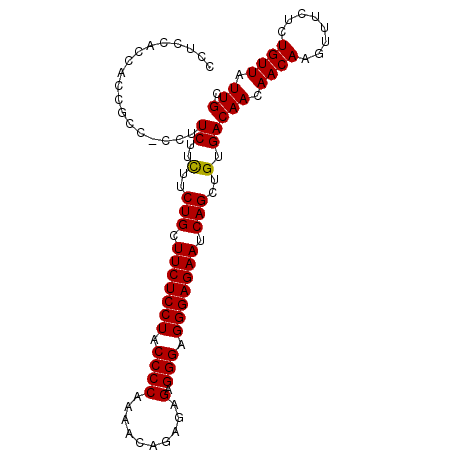
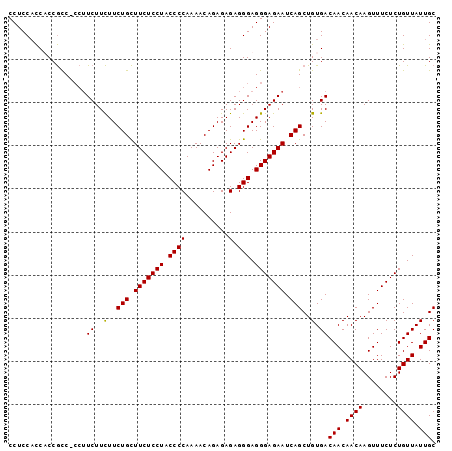
| Location | 13,538,725 – 13,538,823 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13538725 98 - 27905053 GCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCCUCUCUCUGUUUUGGGGUAGGAGAAGCAGAAGAAGAAGG-GACGGUGGUGGAGG ((((((((((......))))))))))(((.((((....(((((.(.(((.((((((((.........))))))))))).).)))-)))))).))).... ( -31.70) >DroSec_CAF1 13583 98 - 1 GCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCCUCUCUCUGUUUUGGGGUAGGAGAAGCAGAAGAAGAAGG-GGCGGAGGUGGAGG ((((((((((......)))))))))).....((...(((((((((.((((((((((((.........))))))).)).))).))-)).)))))...)). ( -31.60) >DroSim_CAF1 10510 99 - 1 GCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCCUCUCUCUGUUUUGGGGUAGGAGAAGCAGAAGAAGAAGGGGGCGGUGGUGGAGG ((((((((((......))))))))))(((.((((...((((((.(.(((.((((((((.........))))))))))).).)))))))))).))).... ( -33.80) >DroYak_CAF1 17408 95 - 1 GCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCCUCUCUCUGUUUUGGGGUAGGAGAAACAGAAAAAGA----GGUGGAGAAGGAGG ((((((((((......)))))))))).....((..((((((...(((((.((((((((.........))))))))...)))----)).))))))..)). ( -31.60) >consensus GCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCCUCUCUCUGUUUUGGGGUAGGAGAAGCAGAAGAAGAAGG_GGCGGAGGUGGAGG ((((((((((......)))))))))).....(((.((((((..((((...........))))..)))))).)))......................... (-26.20 = -26.20 + -0.00)

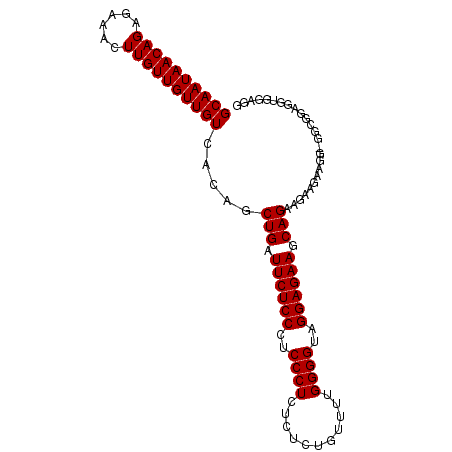
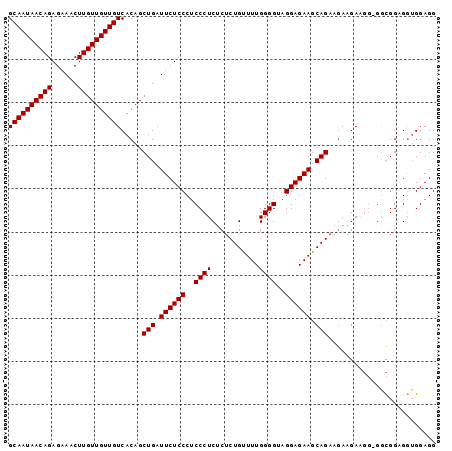
| Location | 13,538,744 – 13,538,858 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -22.20 |
| Energy contribution | -23.44 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13538744 114 - 27905053 CAACAACAAUGGC-----CAAGUAGGCACCGUAAUCAAAGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCC-UCUCUCUGUUUUGGGGUAGGAGAAGCAGAAGAA ...........((-----(.....))).......((...(((((((((((......)))))))))))....(((.((((((..(((-(...........))))..)))))).)))..)). ( -34.60) >DroSec_CAF1 13602 101 - 1 ------------------CAAGUAGGCACCGUAAUCAAAGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCC-UCUCUCUGUUUUGGGGUAGGAGAAGCAGAAGAA ------------------................((...(((((((((((......)))))))))))....(((.((((((..(((-(...........))))..)))))).)))..)). ( -30.30) >DroSim_CAF1 10530 114 - 1 CAACAACAAUGGC-----CAAGUAGGCACCGUAAUCAAAGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCC-UCUCUCUGUUUUGGGGUAGGAGAAGCAGAAGAA ...........((-----(.....))).......((...(((((((((((......)))))))))))....(((.((((((..(((-(...........))))..)))))).)))..)). ( -34.60) >DroYak_CAF1 17424 114 - 1 CAACAACAAUGGA-----CAAGUAGGCACCGUAAUCAAAGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCC-UCUCUCUGUUUUGGGGUAGGAGAAACAGAAAAA ........((((.-----..........)))).......(((((((((((......)))))))))))....(((.((((((..(((-(...........))))..)))))).)))..... ( -31.10) >DroAna_CAF1 11662 104 - 1 AAACAGCCAUUUCCAACGCAAGUAGGCACCGCAAUCACGGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGACUCGCCCUCCCCUC-----GUUCUG--GCAGGAGGA--------- ...((((.....((.((....)).)).............(((((((((((......)))))))))))...))))......(((((((..-----.....)--)..))))).--------- ( -31.20) >consensus CAACAACAAUGGC_____CAAGUAGGCACCGUAAUCAAAGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUCCCUCCC_UCUCUCUGUUUUGGGGUAGGAGAAGCAGAAGAA .......................................(((((((((((......)))))))))))....(((.((((((..(((..............)))..)))))).)))..... (-22.20 = -23.44 + 1.24)

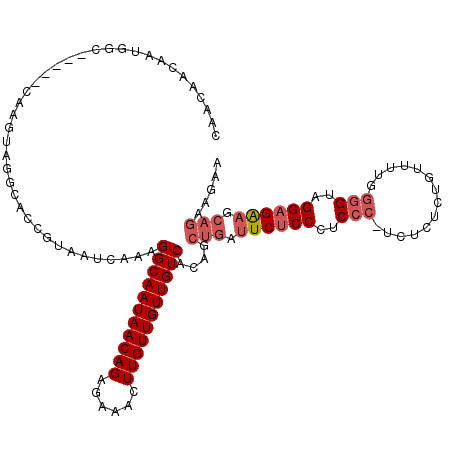
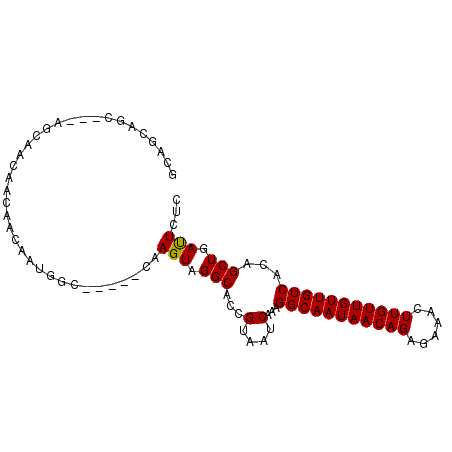
| Location | 13,538,783 – 13,538,874 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -13.69 |
| Energy contribution | -13.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13538783 91 - 27905053 GCAGCAGCAGUAGCAACAACAACAAUGGC-----CAAGUAGGCACCGUAAUCAAAGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUC ....((((.((..(((((((((.....((-----(.....))).((.........))...............)))))))))..)).))))...... ( -24.20) >DroSim_CAF1 10569 88 - 1 GCAGCAGC---AGCAACAACAACAAUGGC-----CAAGUAGGCACCGUAAUCAAAGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUC .......(---(((.............((-----(.....)))............(((((((((((......)))))))))))...))))...... ( -22.70) >DroAna_CAF1 11686 81 - 1 G---------------AAACAGCCAUUUCCAACGCAAGUAGGCACCGCAAUCACGGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGACUCGC (---------------(..((((.....((.((....)).)).............(((((((((((......)))))))))))...))))..)).. ( -22.60) >consensus GCAGCAGC___AGCAACAACAACAAUGGC_____CAAGUAGGCACCGUAAUCAAAGGCAAUAACAGAGAAACUUGUUGUUGUCACAGCUGAUUCUC ....................................(((.(((...(....)...(((((((((((......)))))))))))...))).)))... (-13.69 = -13.47 + -0.22)

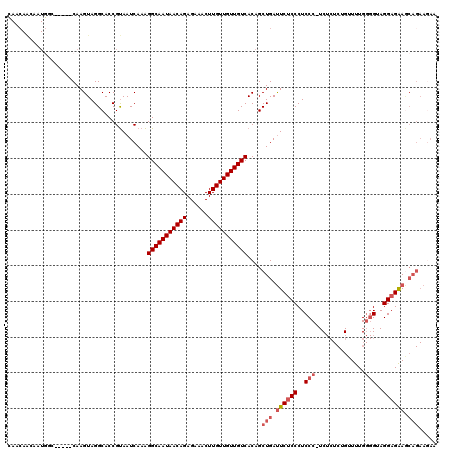
| Location | 13,538,874 – 13,538,982 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.71 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -15.89 |
| Energy contribution | -15.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13538874 108 - 27905053 AAGAAAUCAAAUUGCCGGCAGAGUGUGACUUACGCGUGCAUGCAAUAAAUACAU--------AUACUCGCAUACAUACUUACGUAUCAAACAUACAUCGGUACAACCGAAACGGCA ............(((((...(((((((.....((.(((.(((.........)))--------.))).))....)))))))..((((.....)))).((((.....))))..))))) ( -23.30) >DroSec_CAF1 13703 98 - 1 AAGAAAUCAAAUUGCCGGCAGAGUGUGACUUACGCGUGCAUGCAAUAAAUACAU--------AUACUCGCAUGCAUACUUACGUAUCAAACAUACAUAGGUACAAC---------- ............((((....(.((((....((((.((((((((...........--------......)))))))).....))))....)))).)...))))....---------- ( -19.33) >DroYak_CAF1 17578 112 - 1 AAGAAAUCAAAUUGCCGGCAGAGUGUGACUUACGCGUGCAUGCAAUAAAUACAUAUGUAGAUAUACUCGCAUACAUACUUACGUACCAAGCA----UAGGUACAACCGAAACGGCA ............(((((((.(((((((.((.(((..((.((.......)).))..))))).)))))))))............(((((.....----..)))))........))))) ( -28.10) >consensus AAGAAAUCAAAUUGCCGGCAGAGUGUGACUUACGCGUGCAUGCAAUAAAUACAU________AUACUCGCAUACAUACUUACGUAUCAAACAUACAUAGGUACAACCGAAACGGCA ..........(((((..(((..(((((...))))).)))..)))))....................................(((((...........)))))............. (-15.89 = -15.67 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:39 2006