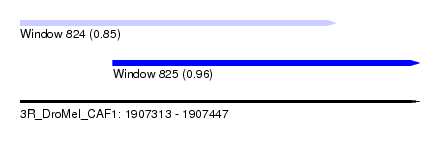
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,907,313 – 1,907,447 |
| Length | 134 |
| Max. P | 0.957778 |

| Location | 1,907,313 – 1,907,419 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.46 |
| Mean single sequence MFE | -24.01 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.850066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1907313 106 + 27905053 CCGAACG-------UGAAAUCCAAAAAUA--UAUCUGAAUUGUCAGGGCCGAA-UGAA-AAAGGAGAC-AAAAAUAAACCGCGAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAA .....((-------((.............--..(((((....)))))......-....-...(....)-..........))))(((((((.((((((....)))))).)))))))... ( -22.10) >DroPse_CAF1 52548 105 + 1 -----AG-------CGGAAUCCAAAAAUAUAUAUUUGCCAUGUCACUGGCAAA-AGGAAAAAGAGGGCAAAAAAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAA -----.(-------(((................(((((((......)))))))-........(....)..........)))).(((((((.((((((....)))))).)))))))... ( -25.10) >DroEre_CAF1 67982 106 + 1 CCAAACG-------UGGAAUCCAAAAAUA--UAUCUGCCUUGUCAGGGGCGAA-UGAA-AAGGGAGAC-AAAAAUAUACCGCCAUUGGUCAUGUUUGAAAGCAAAUGAGGCCAAUUAG ......(-------(((............--(((.((((((....)))))).)-))..-...(....)-.........)))).(((((((.((((((....)))))).)))))))... ( -24.60) >DroYak_CAF1 71832 113 + 1 CCGAACGUGGAACGUGGAAUCCAAAAAUA--UAUCUGCCUUGUCAGGGGCGAA-UGAA-AAAGGAGAC-ACAAAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAA ......((((...(((...(((.......--(((.((((((....)))))).)-))..-...)))..)-)).......)))).(((((((.((((((....)))))).)))))))... ( -27.10) >DroAna_CAF1 53858 107 + 1 CCAAAUA-------UGGAUUCCAAAAAUA--UAUCUGCCAUGUCACAUGCGAAUUGUA-AAAGGAAAC-GAAUAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAAGCCAAUUAA ....(((-------(((............--......)))))).....(((..(((((-...(....)-...)))))..))).((((((..((((((....))))))..))))))... ( -20.07) >DroPer_CAF1 53043 105 + 1 -----AG-------CGGAAUCCAAAAAUAUAUAUUUGCCAUGUCACUGGCAAA-AGGAAAAAGAGGGCAAAAAAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAA -----.(-------(((................(((((((......)))))))-........(....)..........)))).(((((((.((((((....)))))).)))))))... ( -25.10) >consensus CC_AACG_______UGGAAUCCAAAAAUA__UAUCUGCCAUGUCACGGGCGAA_UGAA_AAAGGAGAC_AAAAAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAA ..............(((..................(((((......)))))...........(....)..........)))..(((((((.((((((....)))))).)))))))... (-15.53 = -15.62 + 0.08)

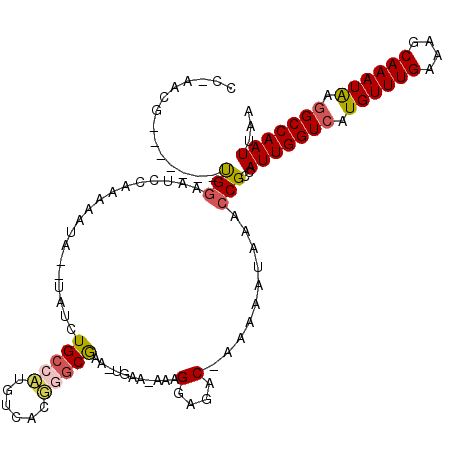
| Location | 1,907,344 – 1,907,447 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.29 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -11.62 |
| Energy contribution | -11.65 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1907344 103 + 27905053 UGUCAGGGCCGAA-UGAA-AAAGGAGAC-AAAAAUAAACCGCGAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAACAAAUGCAAGUACGC-AGCAUAUUUUUGC ((((....((...-....-...)).)))-)..........((((((((((.((((((....)))))).))))))))......(((......))-))).......... ( -22.80) >DroPse_CAF1 52576 99 + 1 UGUCACUGGCAAA-AGGAAAAAGAGGGCAAAAAAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAACAAAUGCAAAUGUGA-AAAAUAU------ ..((((..(((..-...........(((............)))(((((((.((((((....)))))).))))))).......)))....))))-.......------ ( -22.90) >DroEre_CAF1 68013 103 + 1 UGUCAGGGGCGAA-UGAA-AAGGGAGAC-AAAAAUAUACCGCCAUUGGUCAUGUUUGAAAGCAAAUGAGGCCAAUUAGCAAAUGCAAAUACG-GCGCGUAUUUUAGA ..(((........-))).-...(....)-...((.(((((((((((((((.((((((....)))))).)))))))..((....))......)-))).)))).))... ( -24.90) >DroWil_CAF1 64998 93 + 1 UGUCACUGAACCA-UC----------AU-A-AAAAAAAACCCCAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAACAAAUGCAAAUACGC-UGAAUAUUUUCAA ......((((...-..----------..-.-............(((((((.((((((....)))))).)))))))........((......))-........)))). ( -14.30) >DroAna_CAF1 53889 104 + 1 UGUCACAUGCGAAUUGUA-AAAGGAAAC-GAAUAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAAGCCAAUUAACGAAUGCGAACAUCC-CACAUAUUUUUUA ((((.((((((..(((((-...(....)-...)))))..))).((((((..((((((....))))))..))))))......))).).)))...-............. ( -19.60) >DroPer_CAF1 53071 105 + 1 UGUCACUGGCAAA-AGGAAAAAGAGGGCAAAAAAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAACAAAUGCAAAUGUGC-AGAAUAUUUGUAC (((.....)))..-...........(((............)))(((((((.((((((....)))))).))))))).......(((((((((..-...))))))))). ( -23.40) >consensus UGUCACUGGCGAA_UGAA_AAAGGAGAC_AAAAAUAAACCGCCAUUGGUCAUGUUUGAAAGCAAAUAAGGCCAAUUAACAAAUGCAAAUACGC_AGAAUAUUUUUAA ...........................................(((((((.((((((....)))))).)))))))................................ (-11.62 = -11.65 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:01 2006