
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,524,124 – 13,524,424 |
| Length | 300 |
| Max. P | 0.940296 |

| Location | 13,524,124 – 13,524,230 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -51.82 |
| Consensus MFE | -33.99 |
| Energy contribution | -35.15 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.787013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13524124 106 + 27905053 CCACUGGCACCCUG---UCCC------UGCGGCUGGCCGCCUUGGAGCAUUUGGGAUUGCGGCGGCGUGCUAGUGGGUGGACCCAUGUUGGUGGGUCCGUGCGGGGGUCCCAUGU ((((((((((.(((---((((------.((((....))))...)).(((........)))))))).))))))))))..((((((((....))))))))....(((...))).... ( -54.10) >DroSec_CAF1 8723 112 + 1 CCACUGGCACCCUG---UCCCUGACCCUGGGGCUGGCCGCCUUGGAGCAUUUGGGAUUGCGGCGGCGUGCUAGUGGGUGGACCCAUGUUGGUGGGUCCGUGCGGGGGUCCCAUAU ((((((((((...(---((((.......)))))..((((((.....(((........)))))))))))))))))))..((((((((....))))))))....(((...))).... ( -58.00) >DroEre_CAF1 8707 112 + 1 CCACUGGCGCCCUG---UCCCUGGCCCUGGGGCUGGCCGCCCUGAAGCAUUUGCGAUUGCGGCGGCGUGCUAGUGGGUGGACCCAUGUUGGUGGGUCCGUGCGGGGGUCCCAUGU ((((((((((...(---((((.......)))))..(((((((....((....))....).))))))))))))))))..((((((((....))))))))....(((...))).... ( -59.20) >DroWil_CAF1 15730 88 + 1 CCGCUGCCUCCCUGGGCUCCC------------------------AGCAU---GGAUUGUGGCGGUGUGCUCGUAGGUGGACCCAUGCUUGUUGGUCCAUGAGGCGGGCCCAUAU ((((..(.(((((((....))------------------------))...---)))..)..))))((.((((((..(((((((.((....)).)))))))...)))))).))... ( -39.70) >DroYak_CAF1 7996 112 + 1 CCACUGGCGCCCUG---UCCCUGACCCUGGGGCUGGCCGCCCUGGAGCAUUUGCGAUUGUGGCGGCGUGCUAGUGGGUGGACCCAUGUUGGUGGGUCCGUGCGGGGGUCCCAUGU ((((((((((...(---((((.......)))))..(((((((....((....))....).))))))))))))))))..((((((((....))))))))....(((...))).... ( -59.20) >DroAna_CAF1 21678 109 + 1 CCGCCGGUGGUCUG---CCCUUGUCCCUGCCCCUGA---CCCUGUAGCAUCUGCGACUGAGGCGGCGUGCUGGUAGGUGGGCCCAUACUAGUUGGCCCGUGUGGAGGGCCCAUGU ......((((...(---(((((...((((((.....---......(((((((((.......)))).))))))))))).(((((.((....)).))))).....)))))))))).. ( -40.70) >consensus CCACUGGCGCCCUG___UCCCUG_CCCUGGGGCUGGCCGCCCUGGAGCAUUUGCGAUUGCGGCGGCGUGCUAGUGGGUGGACCCAUGUUGGUGGGUCCGUGCGGGGGUCCCAUGU ((((((((((.(((....((........(((((.....)))))...(((........)))))))).))))))))))..((((((((....))))))))(((.((....))))).. (-33.99 = -35.15 + 1.16)

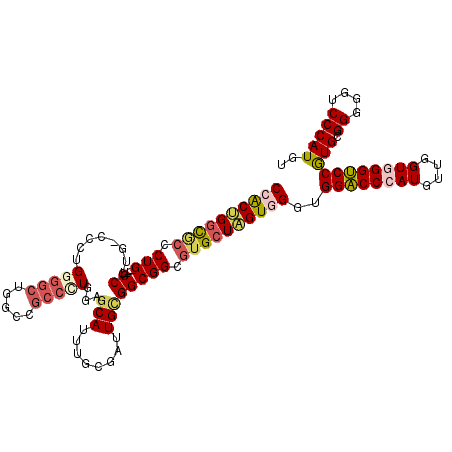
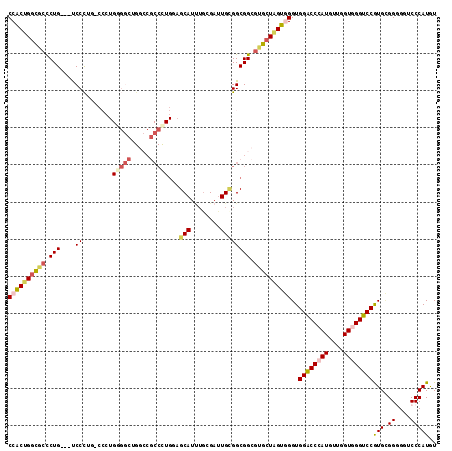
| Location | 13,524,150 – 13,524,270 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.28 |
| Mean single sequence MFE | -39.39 |
| Consensus MFE | -24.27 |
| Energy contribution | -23.88 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13524150 120 - 27905053 ACCGGAAUGCAUCCUGGCAUGCCAAUGGGGCCACCCCACCACAUGGGACCCCCGCACGGACCCACCAACAUGGGUCCACCCACUAGCACGCCGCCGCAAUCCCAAAUGCUCCAAGGCGGC ...((..(((....(((....)))..((((....((((.....))))..))))))).(((((((......)))))))..))........(((((((((........))).....)))))) ( -45.30) >DroVir_CAF1 11617 99 - 1 ACCGGCAUGCAUCCGGCAAUGCCUAUGGGCCCACCACAUCAUAUGGGUCCGCCACAA---------AGUAUGGGGCCACCUACUAGCACGCCGCCUCAGGCU---AUGCUA--------- ...(((((((.....)).)))))...(((((((..........)))))))(((.((.---------....)).))).......(((((.(((......))).---.)))))--------- ( -33.10) >DroPse_CAF1 9016 108 - 1 ACCGGAAUGCACCCUGGCAUGCCAAUGGGUCCGCCCCACCACAUGGGCCCGCCGCACGGGCCAACCAACAUGGGUCCGCCCACGAGCACCCCGCCCCAGUCG---AUGCUC--------- ...((.((((......)))).))..(((((..((((.........((((((.....)))))).........))))..))))).((((((..(......)..)---.)))))--------- ( -38.97) >DroWil_CAF1 15751 107 - 1 ACUGGAAUGCAUCCCGGCAUGCCGAUGGGUCCACCUCAUCAUAUGGGCCCGCCUCAUGGACCAACAAGCAUGGGUCCACCUACGAGCACACCGCCACAAUCC---AUGCU---------- ..((((.((.....(((..(((((..(((((((..........)))))))......((((((..........))))))....)).)))..)))...)).)))---)....---------- ( -33.40) >DroAna_CAF1 21710 117 - 1 ACGGGAAUGCAUCCCGGCAUGCCGAUGGGACCACCUCAUCACAUGGGCCCUCCACACGGGCCAACUAGUAUGGGCCCACCUACCAGCACGCCGCCUCAGUCGCAGAUGCUACAGGG---U ..((((.....))))(((.(((.((((((.....))))))...(((((((.((....))((......))..))))))).......))).)))((((..((.((....)).)).)))---) ( -42.70) >DroPer_CAF1 8237 108 - 1 ACCGGAAUGCACCCUGGCAUGCCAAUGGGUCCGCCCCACCACAUGGGCCCGCCGCACGGGCCCACCAACAUGGGUCCGCCCACGAGCACCCCGCCCCAGUCG---AUGCUC--------- ...((.((((......)))).))..(((((..((((.......((((((((.....)))))))).......))))..))))).((((((..(......)..)---.)))))--------- ( -42.84) >consensus ACCGGAAUGCAUCCUGGCAUGCCAAUGGGUCCACCCCACCACAUGGGCCCGCCGCACGGGCCAACCAACAUGGGUCCACCCACGAGCACGCCGCCCCAGUCG___AUGCUC_________ ..(((..(((.((((((....)))..(((((((..........))))))).......)))..........((((....))))...)))..)))........................... (-24.27 = -23.88 + -0.39)



| Location | 13,524,190 – 13,524,305 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -47.40 |
| Consensus MFE | -25.20 |
| Energy contribution | -26.65 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13524190 115 - 27905053 CGCCGCAGGG---GCCUCAGGGGUACGGCAACGGGCCGACCGGAAUGCAUCCUGGCAUGCCAAUGGGGCCACCCCACCACAUGGGACCCCCGCACGGACCCACCAACAUGGGUCCACC ....((.(((---((((((..(((.((((.....)))))))((.((((......)))).))..)))))))..((((.....))))...)).))..(((((((......)))))))... ( -54.10) >DroVir_CAF1 11645 103 - 1 GUCCACA------GCCGCAGGGAUAUGGCAAUGGGCCGACCGGCAUGCAUCCGGCAAUGCCUAUGGGCCCACCACAUCAUAUGGGUCCGCCACAA---------AGUAUGGGGCCACC .......------(((.((......((((..((((((.(..(((((((.....)).)))))..).))))))(((.......)))....))))...---------....)).))).... ( -37.82) >DroPse_CAF1 9044 118 - 1 CACAGCAGGGCGUGCCGCAAGGCUAUGGCAACGGCCCGACCGGAAUGCACCCUGGCAUGCCAAUGGGUCCGCCCCACCACAUGGGCCCGCCGCACGGGCCAACCAACAUGGGUCCGCC ....((.(((((.(((....)))...(....)((((((...((.((((......)))).))..))))))))))).(((.((((((((((.....))))))......)))))))..)). ( -51.50) >DroWil_CAF1 15778 112 - 1 CGCCCCA------ACCGCAAGGCUACGGCAAUGGACCGACUGGAAUGCAUCCCGGCAUGCCGAUGGGUCCACCUCAUCAUAUGGGCCCGCCUCAUGGACCAACAAGCAUGGGUCCACC .(((...------.((....))....)))..((((((..((((........))))(((((....(((((((..........))))))).((....))........))))))))))).. ( -42.50) >DroYak_CAF1 8068 115 - 1 CGCCUCAGGG---GACUCAAGGCUACGGCAACGGGCCGACCGGUAUGCAUCCUGGUAUGCCAAUGGGGCCACCCCACCACAUGGGACCCCCGCACGGACCCACCAACAUGGGUCCACC .((....(((---(.((((.((((.((....))))))....(((((((......)))))))..(((((...))))).....)))).)))).))..(((((((......)))))))... ( -51.90) >DroAna_CAF1 21747 115 - 1 CGCCACAGGG---ACCUCAAGGAUACGGAAACGGUCCGACGGGAAUGCAUCCCGGCAUGCCGAUGGGACCACCUCAUCACAUGGGCCCUCCACACGGGCCAACUAGUAUGGGCCCACC .......(((---.((....((((.((....))))))..(((((.....))))).(((((.((((((.....)))))).....(((((.......))))).....))))))))))... ( -46.60) >consensus CGCCACAGGG___GCCGCAAGGCUACGGCAACGGGCCGACCGGAAUGCAUCCUGGCAUGCCAAUGGGGCCACCCCACCACAUGGGCCCGCCGCACGGACCAACCAACAUGGGUCCACC ..............((....))....(((...(((((.(..((.((((......)))).))..).)))))..((((.....))))...)))....(((((..........)))))... (-25.20 = -26.65 + 1.45)
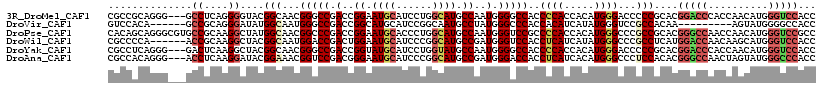


| Location | 13,524,305 – 13,524,424 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13524305 119 - 27905053 ACCGCCACCUCCAGUUGUGGGCGGACCACCACCACCGCAGGGCAGUGGAUCACCGCGUCCACUUAACUAUCUGAAGCAGCAUUUACAGCACAAGGGCGGCUAUGGUGGAAGUCCGACUC- .((((.((.....)).)))).(((((..((((((((((.((((.((((....)))))))).........(((...((..........))...)))))))...))))))..)))))....- ( -42.80) >DroPse_CAF1 9162 116 - 1 ACCGCCGCCUCCGGUGGUGGG---UCCACCGCCUCCUCAAGGCAGUGGCUCGCCACGCCCACUCAACUAUCUGAAGCAGCAUUUACAGCACAAAGGUGGCUACGGUGGCAGCCCCACGC- ...((.(((.(((((((((((---.((((.((((.....)))).))))))))))))(((.(((((......)))....((.......))......)))))..))).))).)).......- ( -47.20) >DroGri_CAF1 11860 111 - 1 ACCACCGC---CGGUG------GGGCCACCGCCACCCCAAGGCAGUGGUUCGCCACGCCCACUCAACUAUCUGAAACAGCAUUUACAACACAAAGGUGGAUACGGCGCCAGUCCAGCUGG .....(((---(((((------....)))).(((((....(((.((((....)))))))...(((......)))....................)))))....))))((((.....)))) ( -37.50) >DroWil_CAF1 15890 116 - 1 ACCACCAC---CCGUUGGACCAGGCCCACCGCCACCUCAGGGCGGUGGAUCACCACGCCCAUUAAACUAUCUUAAGCAGCAUUUACAGCACAAAGGUGGCUACGGCGGCAGUCCUACAC- ........---..((.((((...((((...(((((((..(((((.(((....))))))))..................((.......))....)))))))....).))).)))).))..- ( -40.20) >DroMoj_CAF1 12357 110 - 1 ACCACCAC---CAGUU------GGACCACCACCGCCACAGGGCAGUGGCUCUCCGCGUCCUCUUAACUAUCUUAAGCAGCAUUUACAACACAAAGGUGGAUAUGGCGGUAGUCCAGCGG- .......(---(.(((------((((....(((((((.(((((.((((....))))))))).....(((((((...................)))))))...))))))).)))))))))- ( -37.81) >DroAna_CAF1 21862 119 - 1 UCCACCGCCACCAGUUGUGGGCGGACCUCCUCCACCGCAAGGAAGCGGCUCGCCACGGCCACUGAACUAUUUGAAGCAGCAUUUACAGCACAAAGGCGGCUACGGAGGUAGUCCAACGC- (((((.((.....)).))))).((((..(((((.((....))..(..((.((((.(((...)))..............((.......)).....))))))..))))))..)))).....- ( -42.10) >consensus ACCACCAC___CAGUUGUGGG_GGACCACCGCCACCGCAAGGCAGUGGCUCGCCACGCCCACUCAACUAUCUGAAGCAGCAUUUACAGCACAAAGGUGGCUACGGCGGCAGUCCAACGC_ ......................((((..(((((.......((..((((....))))..))......((((((.....................))))))....)))))..))))...... (-20.94 = -21.50 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:21 2006