
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,515,162 – 13,515,360 |
| Length | 198 |
| Max. P | 0.999821 |

| Location | 13,515,162 – 13,515,280 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -25.64 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13515162 118 + 27905053 GGUUUAGUUAUCAUAUUUACUAUUUUU--AGAUUUGAGUGAAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUUGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGGCG .((((.((((((.......(((....)--))......((((......(((((((....)))))))......)))).((((((((.(((....))).)))))))).))))....)))))). ( -29.80) >DroSec_CAF1 4136 118 + 1 GGUUUAGUUAUCAUAUUUACUAUUUUU--AGAUUUGAGUGAAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGGCG .((((.((((((.......(((....)--))......((((......(((((((....)))))))......)))).((((((((..((....))..)))))))).))))....)))))). ( -27.00) >DroSim_CAF1 4513 118 + 1 GGUUUAGUUAUCAUAUUUACUAUUUUU--AGAUUUGAGUGAAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGGCG .((((.((((((.......(((....)--))......((((......(((((((....)))))))......)))).((((((((..((....))..)))))))).))))....)))))). ( -27.00) >DroEre_CAF1 4589 116 + 1 ----UAGUUAUCAUAUUUACUAUUUUUUAAGAUUUGAGUGAAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGUGAGCCAUUCCAUCGCAGAUACAUUGCAGGCG ----..((.(((.........................((((......(((((((....)))))))......)))).((((((((..((....))..)))))))).))))).......... ( -25.50) >DroYak_CAF1 4866 114 + 1 ----UAGUUAUCAUAUUUACUAUUUUU--AGAUUUGAGUGAAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGACG ----..(((.(((.(((((.......)--)))).)))((((......(((((((....)))))))......)))).((((((((..((....))..))))))))............))). ( -26.40) >consensus GGUUUAGUUAUCAUAUUUACUAUUUUU__AGAUUUGAGUGAAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGGCG ......((.(((.........................((((......(((((((....)))))))......)))).((((((((..((....))..)))))))).))))).......... (-25.64 = -25.64 + 0.00)

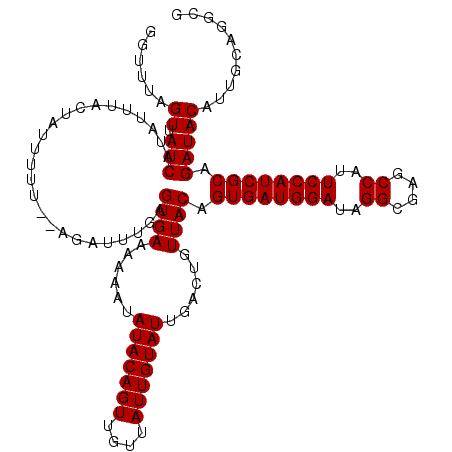
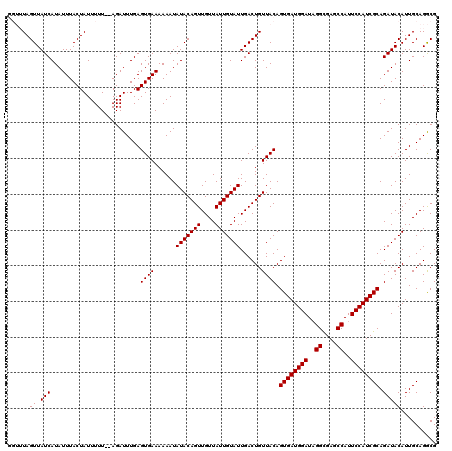
| Location | 13,515,200 – 13,515,320 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.08 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13515200 120 + 27905053 AAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUUGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGGCGCUGCACCUGCUGAGUUCCCGUUUACGUAUCCUUAGAGUUC .......(((((((....))))))).((((......((((((((.(((....))).)))))))).(((((...(((((.......)))))...((........))))))).....)))). ( -35.20) >DroSec_CAF1 4174 120 + 1 AAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGGCGCUGCACCUGCUGAGUUCCCGUUUACGUAUCCUUGGAGUUC .......(((((((....))))))).((((......((((((((..((....))..)))))))).(((((...(((((.......)))))...((........))))))).....)))). ( -32.40) >DroSim_CAF1 4551 120 + 1 AAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGGCGCUGCACCUGCUGAGUUCCCGUUUACGUAUCCUUGGAGUUC .......(((((((....))))))).((((......((((((((..((....))..)))))))).(((((...(((((.......)))))...((........))))))).....)))). ( -32.40) >DroEre_CAF1 4625 120 + 1 AAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGUGAGCCAUUCCAUCGCAGAUACAUUGCAGGCGUUGCACCUGCUGAGUUCCCGUUUACGUAUCCGUGGAGUUC .......(((((((....))))))).((((.((((.((((((((..((....))..)))))))).(((((...(((((.......)))))...((........))))))).)))))))). ( -34.80) >DroYak_CAF1 4900 120 + 1 AAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGACGCUGCAUCUGCUGAGUUCCCUUUUACGUAUCCUUGGAGUUA .......(((((((....)))))))(((((......((((((((..((....))..)))))))).(((((...(((((.......)))))...((........))))))).....))))) ( -31.40) >consensus AAAAAAUAUACAGUUGUUAUUGUAUUGACUGUUACAGUGAUGGAUAGGCGAGCCAUUCCAUCGCAGAUACAUUGCAGGCGCUGCACCUGCUGAGUUCCCGUUUACGUAUCCUUGGAGUUC .......(((((((....))))))).((((......((((((((..((....))..)))))))).(((((...(((((.......)))))...((........))))))).....)))). (-32.40 = -32.08 + -0.32)

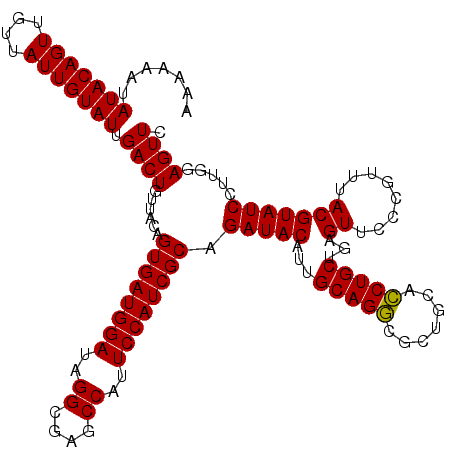
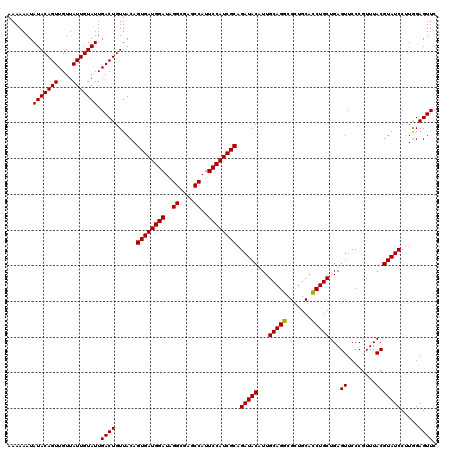
| Location | 13,515,200 – 13,515,320 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -32.68 |
| Energy contribution | -32.36 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13515200 120 - 27905053 GAACUCUAAGGAUACGUAAACGGGAACUCAGCAGGUGCAGCGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCAAUCCAUCACUGUAACAGUCAAUACAAUAACAACUGUAUAUUUUUU .........((((((((....(....)...(((((((...))))))).))))))))..((((((.(((....))).))))))((((...))))..(((((........)))))....... ( -35.60) >DroSec_CAF1 4174 120 - 1 GAACUCCAAGGAUACGUAAACGGGAACUCAGCAGGUGCAGCGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCUAUCCAUCACUGUAACAGUCAAUACAAUAACAACUGUAUAUUUUUU .........((((((((....(....)...(((((((...))))))).))))))))..((((((..((....))..))))))((((...))))..(((((........)))))....... ( -33.00) >DroSim_CAF1 4551 120 - 1 GAACUCCAAGGAUACGUAAACGGGAACUCAGCAGGUGCAGCGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCUAUCCAUCACUGUAACAGUCAAUACAAUAACAACUGUAUAUUUUUU .........((((((((....(....)...(((((((...))))))).))))))))..((((((..((....))..))))))((((...))))..(((((........)))))....... ( -33.00) >DroEre_CAF1 4625 120 - 1 GAACUCCACGGAUACGUAAACGGGAACUCAGCAGGUGCAACGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCACCUAUCCAUCACUGUAACAGUCAAUACAAUAACAACUGUAUAUUUUUU ........(((((((((....(....)...(((((((...))))))).))))))))).((((((..((....))..))))))((((...))))..(((((........)))))....... ( -33.50) >DroYak_CAF1 4900 120 - 1 UAACUCCAAGGAUACGUAAAAGGGAACUCAGCAGAUGCAGCGUCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCUAUCCAUCACUGUAACAGUCAAUACAAUAACAACUGUAUAUUUUUU .........((((((((...((....))..(((((((...))))))).))))))))..((((((..((....))..))))))((((...))))..(((((........)))))....... ( -31.50) >consensus GAACUCCAAGGAUACGUAAACGGGAACUCAGCAGGUGCAGCGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCUAUCCAUCACUGUAACAGUCAAUACAAUAACAACUGUAUAUUUUUU .........((((((((....(....)...(((((((...))))))).))))))))..((((((..((....))..))))))((((...))))..(((((........)))))....... (-32.68 = -32.36 + -0.32)

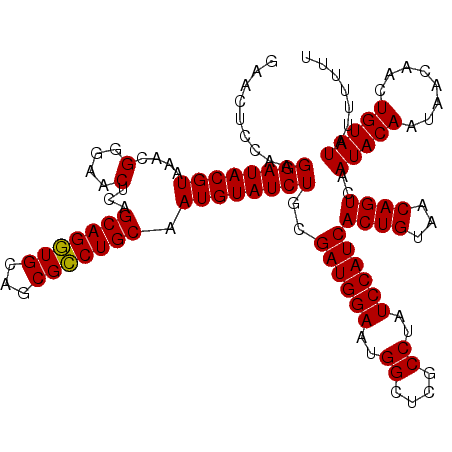
| Location | 13,515,240 – 13,515,360 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -31.54 |
| Energy contribution | -31.78 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13515240 120 - 27905053 GAGAGAGAGAGAGAGUCACGAUGUGCGAGAACUCGUGUCCGAACUCUAAGGAUACGUAAACGGGAACUCAGCAGGUGCAGCGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCAAUCCA .....((((...(.(.(((((.((......))))))).))...))))..((((((((....(....)...(((((((...))))))).))))))))....((((.(((....))).)))) ( -39.30) >DroSec_CAF1 4214 113 - 1 G-------GAGAGAGUCAAGGUGUGCGAGAACUCGUGUCCGAACUCCAAGGAUACGUAAACGGGAACUCAGCAGGUGCAGCGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCUAUCCA (-------((..((((((.((.(..(((....)))..)))....((((.((((((((....(....)...(((((((...))))))).))))))))....)))).)))))).....))). ( -41.80) >DroSim_CAF1 4591 113 - 1 G-------UAGAGAGUCAUGGUGUGCGAGAACUCGUGUCCGAACUCCAAGGAUACGUAAACGGGAACUCAGCAGGUGCAGCGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCUAUCCA (-------(((.(((((((((.(..(((....)))..)))....((((.((((((((....(....)...(((((((...))))))).))))))))....)))))))))))..))))... ( -41.20) >DroEre_CAF1 4665 113 - 1 G-------GAGAAAGUCACGAUGUGCGAGAACACGUGUCCGAACUCCACGGAUACGUAAACGGGAACUCAGCAGGUGCAACGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCACCUAUCCA (-------(((...(.((((.(((......))))))).)....)))).(((((((((....(....)...(((((((...))))))).)))))))))...((((..((....))..)))) ( -38.00) >DroYak_CAF1 4940 113 - 1 G-------GAGAGAUUCACGAUGUGCGAGAACACUUGUCCUAACUCCAAGGAUACGUAAAAGGGAACUCAGCAGAUGCAGCGUCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCUAUCCA .-------...........((((.(((((....(...(((....((...((((((((...((....))..(((((((...))))))).))))))))..)).)))..).))))).)))).. ( -32.50) >consensus G_______GAGAGAGUCACGAUGUGCGAGAACUCGUGUCCGAACUCCAAGGAUACGUAAACGGGAACUCAGCAGGUGCAGCGCCUGCAAUGUAUCUGCGAUGGAAUGGCUCGCCUAUCCA ...................((((.(((((....(((.((((...((...((((((((....(....)...(((((((...))))))).))))))))..))))))))).))))).)))).. (-31.54 = -31.78 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:11 2006