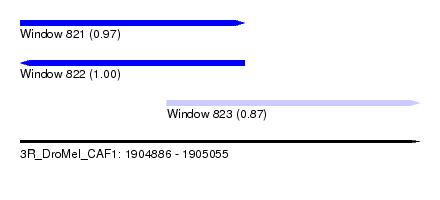
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,904,886 – 1,905,055 |
| Length | 169 |
| Max. P | 0.997523 |

| Location | 1,904,886 – 1,904,981 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
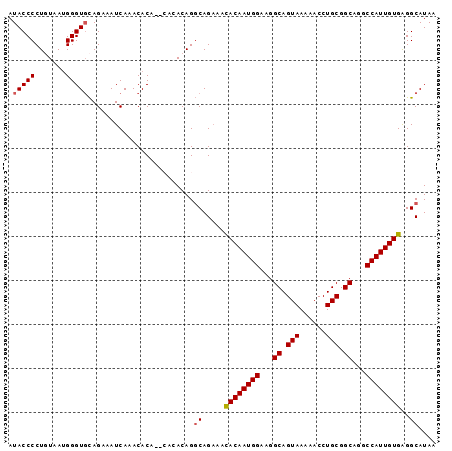
>3R_DroMel_CAF1 1904886 95 + 27905053 UUAUGCCUCACAAUGGCCUGCCGCAGGUUUUUACUGCCUUCCAUUGUGUUUCUGCCUGUGUG--UGUGUUUGAUUUCUGAACCCCUUACAGGGGUAU ..(((((.((((((((......((((.......))))...))))))))......((((((.(--.(.((((.......)))).)).))))))))))) ( -28.20) >DroSec_CAF1 63403 95 + 1 UUAUGCCUCACAAUGGCCUGCCGCAGGUUUUUACUGCCUUCCAUUGUGUUUCUGCCUGUGUG--UGUGUUUGAUUUCUGCACCCAUUACAGGGGUAU ..(((((.((((((((......((((.......))))...))))))))......((((((((--.((((..(....).)))).)).))))))))))) ( -31.60) >DroSim_CAF1 64187 97 + 1 UUAUGCCUCACAAUGGCCUGCCGCAGGUUUUUACUGCCUUCCAUUGUGCUUCUGCCUGUGUGUGUGUGUUUGAUUUCUGCACCCAUUACAGGGGUAU ..(((((.((((((((......((((.......))))...))))))))......((((((.(((.((((..(....).)))).)))))))))))))) ( -31.90) >DroEre_CAF1 65482 93 + 1 UUAUGCCCCACAAUGGCCUGCCGCAGGUUUUUACUGCCUUCCAUUGUAUUUCUGGCUGUG----UGUGUUUGAUUUCUACACCCAUUAGAGGGGUAU ..((((((((((((((......((((.......))))...)))))))...(((((.((.(----((((.........))))).)))))))))))))) ( -31.00) >DroYak_CAF1 69242 91 + 1 UUAUGCCUCACAAUGGCCUGCCGCAGGUUUUUACUGCCUUCCAUUGUAUUGCUGCCUGU------GUGUUUGAUUUCUGCACCCAUUAGAGGGGUAU .(((.(((((((((((......((((.......))))...))))))).........((.------((((..(....).)))).))...)))).))). ( -24.60) >consensus UUAUGCCUCACAAUGGCCUGCCGCAGGUUUUUACUGCCUUCCAUUGUGUUUCUGCCUGUGUG__UGUGUUUGAUUUCUGCACCCAUUACAGGGGUAU ..((((((((((((((......((((.......))))...)))))))..................((((..(....).))))........))))))) (-22.74 = -22.62 + -0.12)

| Location | 1,904,886 – 1,904,981 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
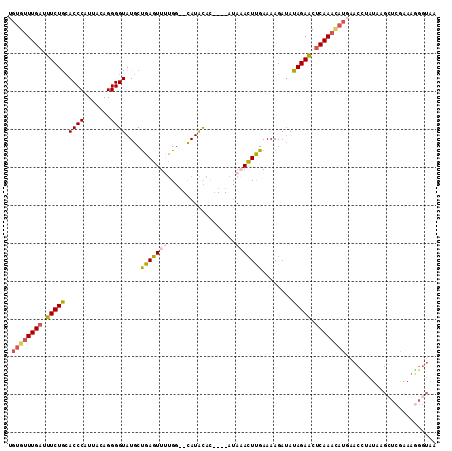
>3R_DroMel_CAF1 1904886 95 - 27905053 AUACCCCUGUAAGGGGUUCAGAAAUCAAACACA--CACACAGGCAGAAACACAAUGGAAGGCAGUAAAAACCUGCGGCAGGCCAUUGUGAGGCAUAA ..(((((.....)))))................--.......((.....((((((((...((.(((......))).))...))))))))..)).... ( -26.10) >DroSec_CAF1 63403 95 - 1 AUACCCCUGUAAUGGGUGCAGAAAUCAAACACA--CACACAGGCAGAAACACAAUGGAAGGCAGUAAAAACCUGCGGCAGGCCAUUGUGAGGCAUAA ...(((((((..((.(((...........))).--)).)))))......((((((((...((.(((......))).))...)))))))).))..... ( -24.90) >DroSim_CAF1 64187 97 - 1 AUACCCCUGUAAUGGGUGCAGAAAUCAAACACACACACACAGGCAGAAGCACAAUGGAAGGCAGUAAAAACCUGCGGCAGGCCAUUGUGAGGCAUAA .....(((((..((.(((...........))).))...)))))..(...((((((((...((.(((......))).))...))))))))...).... ( -24.90) >DroEre_CAF1 65482 93 - 1 AUACCCCUCUAAUGGGUGUAGAAAUCAAACACA----CACAGCCAGAAAUACAAUGGAAGGCAGUAAAAACCUGCGGCAGGCCAUUGUGGGGCAUAA .((((((......))).))).............----....(((.....((((((((...((.(((......))).))...)))))))).))).... ( -27.40) >DroYak_CAF1 69242 91 - 1 AUACCCCUCUAAUGGGUGCAGAAAUCAAACAC------ACAGGCAGCAAUACAAUGGAAGGCAGUAAAAACCUGCGGCAGGCCAUUGUGAGGCAUAA .(((((.......)))))..............------.......((..((((((((...((.(((......))).))...))))))))..)).... ( -23.80) >consensus AUACCCCUGUAAUGGGUGCAGAAAUCAAACACA__CACACAGGCAGAAACACAAUGGAAGGCAGUAAAAACCUGCGGCAGGCCAUUGUGAGGCAUAA .(((((.......)))))........................((.....((((((((...((.(((......))).))...))))))))..)).... (-21.62 = -21.78 + 0.16)

| Location | 1,904,948 – 1,905,055 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -15.89 |
| Energy contribution | -16.61 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
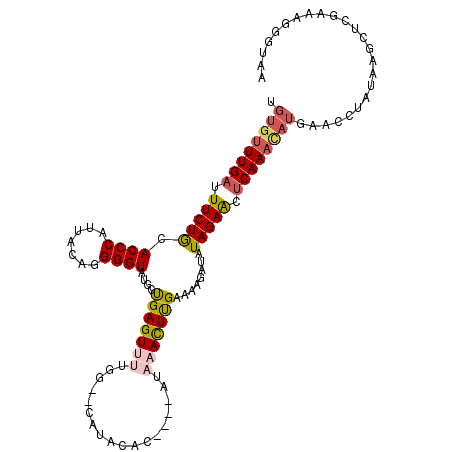
>3R_DroMel_CAF1 1904948 107 + 27905053 UGUGUUUGAUUUCUGAACCCCUUACAGGGGUAUGCUGAGUUUUGG--CAUACAC----AUAAACUUGAAAAGAUAUAGAACACAAAUAUGAACUUAUAAGCUCGAAAAGGUAA (((((((....(((....(((.....)))((((((..(...)..)--)))))..----............)))....)))))))....(((.((....)).)))......... ( -23.10) >DroSec_CAF1 63465 107 + 1 UGUGUUUGAUUUCUGCACCCAUUACAGGGGUAUGCUGAGUUGUGG--CAUACAU----AUAAACUCGAAAAGAUAUAGAACUCAAACAUGAACCUAUAAGCUCGAAAGGGUAA .((((((((.(((((..(((......)))(((((((.......))--)))))..----.................))))).))))))))..........((((....)))).. ( -32.20) >DroSim_CAF1 64251 107 + 1 UGUGUUUGAUUUCUGCACCCAUUACAGGGGUAUGCUGAGUUUUGG--CAUACAU----AUAAACUCGAAAAGAUAUAGAACUCAAACAUGAACCUAUAAGCUCGAAAGGGUAA .((((((((.(((((..(((......)))((((((..(...)..)--)))))..----.................))))).))))))))..........((((....)))).. ( -32.60) >DroEre_CAF1 65542 87 + 1 UGUGUUUGAUUUCUACACCCAUUAGAGGGGUAUGCUGAGUUUGGGA-CAUGUAC----ACAUACUUGAAAAGAAAUAGAGCUCAA---------------------AGGGUAG ((.((((.((((((..((((.......)))).....((((.(((..-......)----.)).))))....)))))).)))).)).---------------------....... ( -16.40) >DroYak_CAF1 69301 112 + 1 -GUGUUUGAUUUCUGCACCCAUUAGAGGGGUAUGCCGAGUUCUGAAGUAUAUACAUAUAUAUAUUUGAAAAGAUAUAGAGCUCAAACAUUAACUUAUAUGAUCAAAAGGGUAG -............(((.(((......))))))((((((((((((..(((((....))))).(((((....)))))))))))))...(((........)))........)))). ( -23.80) >consensus UGUGUUUGAUUUCUGCACCCAUUACAGGGGUAUGCUGAGUUUUGG__CAUACAC____AUAAACUUGAAAAGAUAUAGAACUCAAACAUGAACCUAUAAGCUCGAAAGGGUAA .((((((((.(((((.((((.......))))....(((((((..................)))))))........))))).))))))))........................ (-15.89 = -16.61 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:59 2006