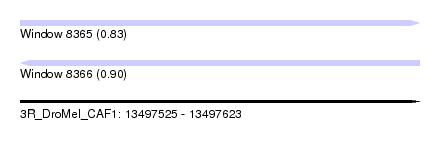
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,497,525 – 13,497,623 |
| Length | 98 |
| Max. P | 0.897285 |

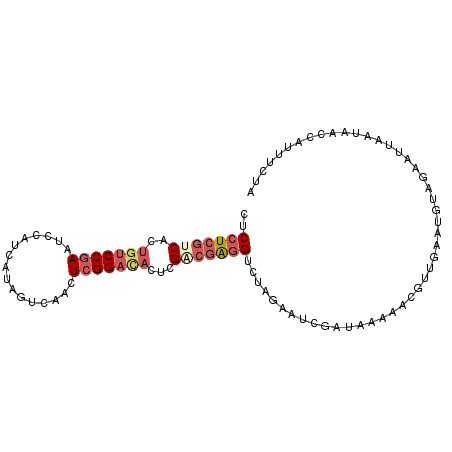
| Location | 13,497,525 – 13,497,623 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13497525 98 + 27905053 UAGAAAUGUUCAUUACUUCUACAUACAACGUUUUUAUCGAUUCUAGACCUCGCCGAGUGUCCGAGUUGACUAUGAUGGAUUCGGACAGUGACGAGGAG ((((((((((................))))))))))...........(((((.((..((((((((((.(......).)))))))))).)).))))).. ( -27.79) >DroSec_CAF1 47433 98 + 1 UAGGGAUGGUUAUUAAUGCUACAUUCAACGUUUUUAUCAAUUCUAGACCUCGCCGAGUGUCCGAGUUGACUAUGAUGGAUUCGGACAGUGACGAGGAG (((((.((((....((((..........))))...)))).)))))..(((((.((..((((((((((.(......).)))))))))).)).))))).. ( -28.30) >DroSim_CAF1 46389 98 + 1 UAGAAAUGGUUAUUAAUUCUAUAUUCAACGUUUUUAUCGAUUCUAGACCUCGUCGAGUGUCCGAGUUGACUAUGAUGGAUUCGGACAGCGACGAGGAG ((((((((....................))))))))...........((((((((..((((((((((.(......).)))))))))).)))))))).. ( -33.45) >DroEre_CAF1 44525 98 + 1 UAGAAAUGGUUAUUAAUUCUACAGUCAACGUUUUUAUUGAUUCUAGACCUCGGCGAGUCUCCGAGUUGACUAUGAUGGAUUCGGACAGUGACGAGGAG .................((((.((((((........)))))).))))(((((.((....((((((((.(......).))))))))...)).))))).. ( -24.90) >DroYak_CAF1 54072 87 + 1 UAAAAAUGGGAAUUAAUUA-----------AUUCUAUCGAUUCUAGACCUCGUCGAGUAUCCGAGUUGACUAUGAUGGAUUUGGACAGUGACGAGGAG ........(((((..((..-----------.....))..)))))...((((((((....((((((((.(......).))))))))...)))))))).. ( -25.20) >DroMoj_CAF1 50113 91 + 1 ---GAAGGCAAU----GUCUGUAUUUCAGAUCAUUUCCGCUUACAGUCCCAGACGCCGAGCCGAGUUGACAAUGAUGGACUCGGACAGUGAUGAUGAG ---...((((((----(((((.....)))).))))..........(((...))))))...(((((((.(......).))))))).((....))..... ( -22.50) >consensus UAGAAAUGGUUAUUAAUUCUACAUUCAACGUUUUUAUCGAUUCUAGACCUCGCCGAGUGUCCGAGUUGACUAUGAUGGAUUCGGACAGUGACGAGGAG ...............................................(((((.((....((((((((.(......).))))))))...)).))))).. (-16.40 = -16.52 + 0.11)

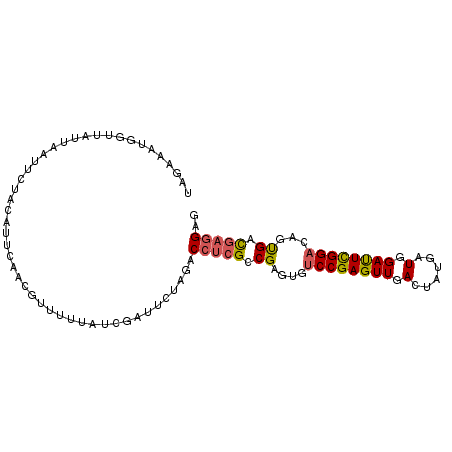
| Location | 13,497,525 – 13,497,623 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -21.79 |
| Consensus MFE | -13.49 |
| Energy contribution | -14.57 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13497525 98 - 27905053 CUCCUCGUCACUGUCCGAAUCCAUCAUAGUCAACUCGGACACUCGGCGAGGUCUAGAAUCGAUAAAAACGUUGUAUGUAGAAGUAAUGAACAUUUCUA ..(((((((..(((((((................)))))))...)))))))((((.(..((((......))))..).))))................. ( -23.79) >DroSec_CAF1 47433 98 - 1 CUCCUCGUCACUGUCCGAAUCCAUCAUAGUCAACUCGGACACUCGGCGAGGUCUAGAAUUGAUAAAAACGUUGAAUGUAGCAUUAAUAACCAUCCCUA ..(((((((..(((((((................)))))))...))))))).(((.(.(..((......))..).).))).................. ( -21.99) >DroSim_CAF1 46389 98 - 1 CUCCUCGUCGCUGUCCGAAUCCAUCAUAGUCAACUCGGACACUCGACGAGGUCUAGAAUCGAUAAAAACGUUGAAUAUAGAAUUAAUAACCAUUUCUA ..((((((((.(((((((................)))))))..))))))))((((...(((((......)))))...))))................. ( -28.59) >DroEre_CAF1 44525 98 - 1 CUCCUCGUCACUGUCCGAAUCCAUCAUAGUCAACUCGGAGACUCGCCGAGGUCUAGAAUCAAUAAAAACGUUGACUGUAGAAUUAAUAACCAUUUCUA ..(((((.(..(.(((((................))))).)...).)))))((((.(.(((((......))))).).))))................. ( -18.59) >DroYak_CAF1 54072 87 - 1 CUCCUCGUCACUGUCCAAAUCCAUCAUAGUCAACUCGGAUACUCGACGAGGUCUAGAAUCGAUAGAAU-----------UAAUUAAUUCCCAUUUUUA ..(((((((..(((((....................)))))...)))))))....((((..((.....-----------..))..))))......... ( -16.45) >DroMoj_CAF1 50113 91 - 1 CUCAUCAUCACUGUCCGAGUCCAUCAUUGUCAACUCGGCUCGGCGUCUGGGACUGUAAGCGGAAAUGAUCUGAAAUACAGAC----AUUGCCUUC--- ((((....(.(((.((((((.((....))...))))))..))).)..)))).........((.((((.((((.....)))))----))).))...--- ( -21.30) >consensus CUCCUCGUCACUGUCCGAAUCCAUCAUAGUCAACUCGGACACUCGACGAGGUCUAGAAUCGAUAAAAACGUUGAAUGUAGAAUUAAUAACCAUUUCUA ..(((((((..(((((((................)))))))...)))))))............................................... (-13.49 = -14.57 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:58 2006