
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,484,666 – 13,484,843 |
| Length | 177 |
| Max. P | 0.970494 |

| Location | 13,484,666 – 13,484,767 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13484666 101 + 27905053 UUGAUUUGUUUGGUCUUCAACUCAGCUCAACUCAACCCCAGCAGCAUCAACAAUAACACGUCUCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGAGGUC ((((((((((.((.......................)).))))).))))).......((.(((.(((...(((((((....))))))).)))..))).)). ( -24.00) >DroSec_CAF1 34611 100 + 1 UUGAUUUGUUUGGUC-CCAACUCAGCUCAACUCAACCCCACCAGCAUCAACAAUAACACGUCUCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGAGGAC ((((.(((.(((...-.)))..))).)))).............................(((..(((.(((((((((....)))))))...)))))..))) ( -24.70) >DroSim_CAF1 33479 101 + 1 UUGAGUUGUUUGGUCCCCAACUCAGCUCAACUCAACCCCAGCAGCAUCAACAAUAACACGUCUCCUUAUCGGAACCGGAAACGGCUCCCAAGAAAGAGGUC ((((((((.((((...))))..))))))))...........................((.(((.(((...(((.(((....))).))).)))..))).)). ( -25.40) >DroEre_CAF1 31224 101 + 1 UUGAUUUGCUUGGUCCCCAACUCAACUCAACUCAACCCCCGCAGGAUCAACAAUAACACGUUCCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGAGGUC ((((.(((.((((...))))..))).))))(((.........(((...(((........))).)))....(((((((....))))))).......)))... ( -24.20) >DroYak_CAF1 39278 90 + 1 UUGAUUUGUUUG-UCCCCAA----------CUCAACCCCCGCAGGAUCAACAAUAACACGACUCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGGGGUC .....(((((.(-(((....----------.............)))).)))))......((((((((.(((((((((....)))))))...)))))))))) ( -31.13) >consensus UUGAUUUGUUUGGUCCCCAACUCAGCUCAACUCAACCCCAGCAGCAUCAACAAUAACACGUCUCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGAGGUC ((((((((((.((.......................)).))))).))))).............((((.(((((((((....)))))))...))..)))).. (-19.02 = -19.94 + 0.92)

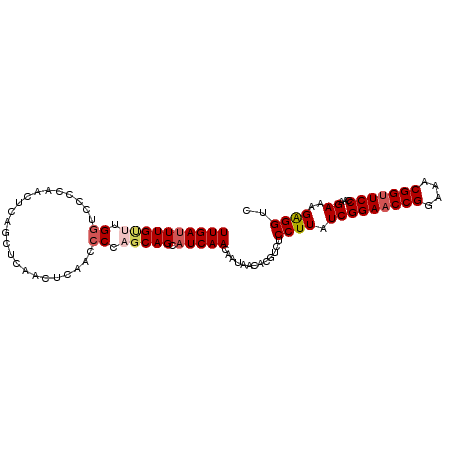

| Location | 13,484,703 – 13,484,806 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -37.51 |
| Consensus MFE | -22.92 |
| Energy contribution | -24.53 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13484703 103 + 27905053 CCAGCAGCAUCAACAAUAACACGU-------------CUCCUUAUCGGAACCGGAAACGGUUCCCAA--GAAAGAGGUCCUAAAAGAGUCCUCA-AAGUCACGGUGGCGUGUGUGACAA .............((...((((((-------------(.((.....(((((((....)))))))...--....((((.((.....).).)))).-.......)).))))))).)).... ( -36.60) >DroPse_CAF1 45156 115 + 1 -CAGCAAGAACAACAAUAGCACGUUCACGUUCCUGUGCCCGUUAUCGGAACCGGAAACGGGUCCACACAGAACGA---AAGGAAAAAGUCCACAAAAGUCACGGUGGCGUGUGUGACAA -......((((...........)))).(((((.((((((((((.(((....))).))))))....))))))))).---.........(((((((...(((.....))).)))).))).. ( -35.90) >DroSec_CAF1 34647 103 + 1 CCACCAGCAUCAACAAUAACACGU-------------CUCCUUAUCGGAACCGGAAACGGUUCCCAA--GAAAGAGGACCUGAAAGAGUCCUCA-AAGUCACGGUGGCGUGUGUGACAA .............((...((((((-------------(.((.....(((((((....)))))))...--....((((((.(....).)))))).-.......)).))))))).)).... ( -41.00) >DroEre_CAF1 31261 103 + 1 CCCGCAGGAUCAACAAUAACACGU-------------UCCCUUAUCGGAACCGGAAACGGUUCCCAA--GAAAGAGGUCCUGAAAGAGUCCUCA-AAGUCACGGUGGCGUGUGUGACAA .............((...((((((-------------..((.....(((((((....)))))))...--....((((.(.(....).).)))).-.......))..)))))).)).... ( -36.20) >DroYak_CAF1 39304 103 + 1 CCCGCAGGAUCAACAAUAACACGA-------------CUCCUUAUCGGAACCGGAAACGGUUCCCAA--GAAAGGGGUCCUGAAGGAGUCCUCA-AAGUCACGGUGGCGUGUGUGACAA ...(.(((((.........((.((-------------((((((.(((((((((....)))))))...--)))))))))).)).....)))))).-..((((((........)))))).. ( -37.24) >DroPer_CAF1 42256 115 + 1 -CAGCAAGAACAACAAUAACACGUUCACGUUCCUGUGCCCGUUAUCGGAACCGGAAACGGGUCCACACAGAACGA---AAGGAAAAAGUCCACAAAAGUCACGCUGGCGUGUGUGACAA -......((((...........)))).(((((.((((((((((.(((....))).))))))....))))))))).---..(((.....)))......(((((((......))))))).. ( -38.10) >consensus CCAGCAGGAUCAACAAUAACACGU_____________CCCCUUAUCGGAACCGGAAACGGUUCCCAA__GAAAGAGGUCCUGAAAGAGUCCUCA_AAGUCACGGUGGCGUGUGUGACAA .............((...((((((......................(((((((....))))))).........((((.(........).)))).............)))))).)).... (-22.92 = -24.53 + 1.61)



| Location | 13,484,730 – 13,484,843 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13484730 113 + 27905053 CUUAUCG-GAACCGGAAACGGUUCCCAA--GAAAGAGGUCCUAAAAGAGUCCUCA-AAGUCACGGUGGCGUGUGUGACAAGUCCUUUCUAAUGAGCAGGAAGAAUGCACGUA---GACUG ......(-((((((....)))))))...--....((((.((.....).).)))).-......((((.((((((((......(((((((....))).))))...)))))))).---.)))) ( -39.70) >DroPse_CAF1 45195 102 + 1 GUUAUCG-GAACCGGAAACGGGUCCACACAGAACGA---AAGGAAAAAGUCCACAAAAGUCACGGUGGCGUGUGUGACAAGUCCUUUCUAAUGAGCAGGAA--------------GAGGG (((...(-((.(((....))).))).........((---(((((....(((((((...(((.....))).)))).)))...))))))).....))).....--------------..... ( -33.60) >DroSec_CAF1 34674 113 + 1 CUUAUCG-GAACCGGAAACGGUUCCCAA--GAAAGAGGACCUGAAAGAGUCCUCA-AAGUCACGGUGGCGUGUGUGACAAGUCCUUUCUAAUGAGCAGGAAGAAUGCACGGA---GACUG .((((((-((((((....)))))))...--....((((((.(....).)))))).-.......)))))(((((((......(((((((....))).))))...)))))))..---..... ( -41.10) >DroSim_CAF1 33543 113 + 1 CUUAUCG-GAACCGGAAACGGCUCCCAA--GAAAGAGGUCCUGAAAGAGUCCUCA-AAGUCACGGUGGCGUGUGUGACAAGUCCUUUGUAAUGAGCAGGAAGAAUGCACGUA---GACUG ......(-((.(((....))).)))...--....((((.(.(....).).)))).-......((((.((((((((......((((..((.....))))))...)))))))).---.)))) ( -33.20) >DroAna_CAF1 35744 94 + 1 CUUAUCGGGAACCGGAAACGG----------------------AAAAAGUCCACA-AAGUCACGGUGGCGUAUGUGACAAGUCCUUUCUAAUGAGCAGGAAGAAUGCACGUU---UACAA ......((((.(((....)))----------------------............-..((((((........))))))...))))....((((.(((.......))).))))---..... ( -22.00) >DroPer_CAF1 42295 116 + 1 GUUAUCG-GAACCGGAAACGGGUCCACACAGAACGA---AAGGAAAAAGUCCACAAAAGUCACGCUGGCGUGUGUGACAAGUCCUUUCUAAUGAGCAGGAAGAAGGCGAGCAUAAGAGGG ((..(((-((.(((....))).))).........((---(((((....(((((((...(((.....))).)))).)))...))))))).............))..))............. ( -35.70) >consensus CUUAUCG_GAACCGGAAACGGGUCCCAA__GAAAGA___CCUGAAAAAGUCCACA_AAGUCACGGUGGCGUGUGUGACAAGUCCUUUCUAAUGAGCAGGAAGAAUGCACGUA___GACUG ...........(((....))).....................................((((((........))))))...(((((((....))).)))).................... (-18.20 = -18.37 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:51 2006