| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,900,747 – 1,900,849 |
| Length | 102 |
| Max. P | 0.596586 |

| Location | 1,900,747 – 1,900,849 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -20.18 |
| Energy contribution | -19.55 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
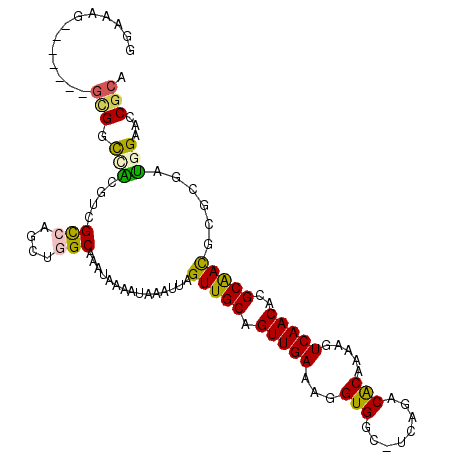
>3R_DroMel_CAF1 1900747 102 - 27905053 GGCAAG-------GCGGCCAGGUCGCCAGCUGGCAAAUAAAAUAAAUUAGUUGCAGUUGAAAGGUGGC-UCAGACACAAAAGUCAACACGCAACGCGCGAUGGAACCGCA .((..(-------(((((...))))))......................(((((.(((((...(((..-.....))).....)))))..)))))))(((.......))). ( -30.10) >DroVir_CAF1 67458 101 - 1 GGAAGG-------GCGUCUGCGUCGCCUGCUUGCAAAUAAAAUAAAUUAGUUGCAGUUGAAAGGUGGC-UCAGACGCAAAAGUCAACACGCGAUGU-CGUCGGUGCCGCA ....((-------(((((((.(((((((..((((((.(((......))).)))))).....)))))))-.))))))).........(((.((((..-.)))))))))... ( -36.90) >DroEre_CAF1 60965 102 - 1 GGGGAG-------GCGGCCAGGUCGCCAGCUGGCAAAUAAAAUAAAUUAGUUGCAGUUGAAAGGUGGC-UCAGACACAAAAGUCAACACGCAACGCGCGAUGGAACCGCA ......-------(((((((.((.(((....)))...............(((((.(((((...(((..-.....))).....)))))..)))))..))..)))..)))). ( -29.40) >DroWil_CAF1 54264 103 - 1 GGUUUG-------GUGGUGGUGGUGCCAGCUGGCAAAUAAAAUAAAUUAGUUGCAGUUGAAAGGUGGCUUCAGACACAAAAGUCAACACGCAACGCGCGAUGGAACCGCA (((((.-------((.(((....((((....))))..............(((((.(((((...(((........))).....)))))..))))).))).)).)))))... ( -33.70) >DroMoj_CAF1 55870 108 - 1 GCAGUGGAUGUUCGCGUCUGCGUCGCCUGCUUGCAAAUAAAAUAAAUUAGUUGCAGUUGAAAGGUGGC-UCAGACGCAAAAGUCAACACGCGAUGU-CGUCGGCACCGCA ...((((((.((.(((((((.(((((((..((((((.(((......))).)))))).....)))))))-.))))))).)).)))..)))(((.(((-.....))).))). ( -41.10) >DroAna_CAF1 48031 94 - 1 ---------------GCCAAUGCCGUCAGCUGGCAAAUAAAAUAAAUUAGUUGCAGUUGAAAGGUGGC-UCAGACACAAAAGUCAACACGCAACGCGCGAUGGAACCGCA ---------------((...(((((.....)))))..............(((((.(((((...(((..-.....))).....)))))..)))))))(((.......))). ( -25.40) >consensus GGAAAG_______GCGGCCACGUCGCCAGCUGGCAAAUAAAAUAAAUUAGUUGCAGUUGAAAGGUGGC_UCAGACACAAAAGUCAACACGCAACGCGCGAUGGAACCGCA .............(((.(((....(((....)))...............(((((.(((((...(((........))).....)))))..)))))......)))...))). (-20.18 = -19.55 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:55 2006