
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,439,309 – 13,439,399 |
| Length | 90 |
| Max. P | 0.596874 |

| Location | 13,439,309 – 13,439,399 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -16.53 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.17 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13439309 90 + 27905053 U--UUUU--ACAUUUCUUUUCUCAUGCC-CAACUU-UACUCUUGUUGCUCUCAAAAAACCUGAAAUUUUCAGGGUCAAAUCGCAUGCAUAAGGCAA .--....--......((((...(((((.-((((..-.......))))...........(((((.....)))))........)))))...))))... ( -15.20) >DroPse_CAF1 20812 91 + 1 AUUUUGU--UCAUUUCUUUUCUCAUGCC-CAAUUU--UGUGUGUGUGCUCUCAAAAAACCUGAAAUUUUCAGGGGCAAAUCAUGUGCAUAAGGCAA .......--...............((((-.....(--((((..((((...........(((((.....))))).......))))..))))))))). ( -17.57) >DroSim_CAF1 8622 90 + 1 U--UUUU--ACAUUUCUUUUCUCAUGCC-CAACUU-UACCCUUGUUGCUCUCAAAAAACCUGAAAUUUUCAGGGGCAAAUCGCAUGCAUAAGGCAA .--....--...............((((-((((..-.......))))...........(((((.....))))))))).......(((.....))). ( -15.40) >DroYak_CAF1 6688 90 + 1 U--UUUU--ACAUUUCUUUUCUCAUGCC-CAACUU-UACUCUUGUUGCUCUCAAAAAACCUGAAAUUUUCAGGGGCAAAUCGCAUGCAUAAGGCAU .--....--...............((((-((((..-.......))))...........(((((.....)))))))))......((((.....)))) ( -15.70) >DroAna_CAF1 8028 93 + 1 U--UUUUUCCCAUUUCUUUUCUCAUGCC-CAAUUUUUUCUGUUUUUGCUCUCAAAAAACCUGAAAUUUUCAGGGGCAAAUCAUGUGCAUAAGGCAA .--.....................((((-(..........((((((.......))))))((((.....))))))))).......(((.....))). ( -16.60) >DroPer_CAF1 14279 91 + 1 NNNNNNN--NNNNNNNNUUUCUCAUGCCCCAAUUU--UGUGUGUAUGCUCUCAAA-AACCUGAAAUUUUCAGGGGCAAAUCAUGUGCAUAAGGCAA .......--...............((((......(--((((..((((........-..(((((.....))))).......))))..))))))))). ( -18.73) >consensus U__UUUU__ACAUUUCUUUUCUCAUGCC_CAACUU_UACUCUUGUUGCUCUCAAAAAACCUGAAAUUUUCAGGGGCAAAUCACAUGCAUAAGGCAA ........................((((..............................(((((.....))))).(((.......)))....)))). (-12.00 = -12.17 + 0.17)



| Location | 13,439,309 – 13,439,399 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -15.88 |
| Energy contribution | -15.38 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13439309 90 - 27905053 UUGCCUUAUGCAUGCGAUUUGACCCUGAAAAUUUCAGGUUUUUUGAGAGCAACAAGAGUA-AAGUUG-GGCAUGAGAAAAGAAAUGU--AAAA--A ....(((((((...((((((.(((((((.....)))))..(((((.......))))))).-))))))-.)))))))...........--....--. ( -19.40) >DroPse_CAF1 20812 91 - 1 UUGCCUUAUGCACAUGAUUUGCCCCUGAAAAUUUCAGGUUUUUUGAGAGCACACACACA--AAAUUG-GGCAUGAGAAAAGAAAUGA--ACAAAAU ....(((((((.((.........(((((.....)))))..(((((.(........).))--))).))-.)))))))...........--....... ( -15.50) >DroSim_CAF1 8622 90 - 1 UUGCCUUAUGCAUGCGAUUUGCCCCUGAAAAUUUCAGGUUUUUUGAGAGCAACAAGGGUA-AAGUUG-GGCAUGAGAAAAGAAAUGU--AAAA--A ....(((((((...((((((..((((.....((((((.....))))))......))))..-))))))-.)))))))...........--....--. ( -23.30) >DroYak_CAF1 6688 90 - 1 AUGCCUUAUGCAUGCGAUUUGCCCCUGAAAAUUUCAGGUUUUUUGAGAGCAACAAGAGUA-AAGUUG-GGCAUGAGAAAAGAAAUGU--AAAA--A ....(((((((...((((((...(((((.....))))).((((((.......))))))..-))))))-.)))))))...........--....--. ( -18.80) >DroAna_CAF1 8028 93 - 1 UUGCCUUAUGCACAUGAUUUGCCCCUGAAAAUUUCAGGUUUUUUGAGAGCAAAAACAGAAAAAAUUG-GGCAUGAGAAAAGAAAUGGGAAAAA--A ....(((((((...((((((...(((((.....))))).((((((....))))))......))))))-.))))))).................--. ( -18.10) >DroPer_CAF1 14279 91 - 1 UUGCCUUAUGCACAUGAUUUGCCCCUGAAAAUUUCAGGUU-UUUGAGAGCAUACACACA--AAAUUGGGGCAUGAGAAANNNNNNNN--NNNNNNN ....(((((((.((.....))(((((((.....))))(((-(....)))).........--.....))))))))))...........--....... ( -16.30) >consensus UUGCCUUAUGCACACGAUUUGCCCCUGAAAAUUUCAGGUUUUUUGAGAGCAAAAACAGAA_AAAUUG_GGCAUGAGAAAAGAAAUGU__AAAA__A ....(((((((...((((((...(((((.....))))).....((....))..........))))))..))))))).................... (-15.88 = -15.38 + -0.50)

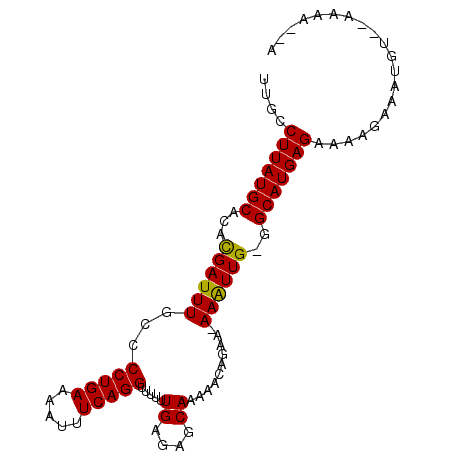

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:07 2006