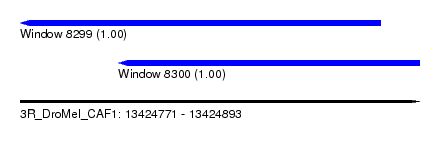
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,424,771 – 13,424,893 |
| Length | 122 |
| Max. P | 0.999932 |

| Location | 13,424,771 – 13,424,881 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.80 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -14.06 |
| Energy contribution | -15.42 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13424771 110 - 27905053 UCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGGGCCAACACCACAGUUGGUCCGCACCC----CUCGAUUC------CGAUUCCACUCC .....((((((((..(((.................((((((((....)))))))).......((((((((......)))))))))))...----....))))------))))........ ( -33.10) >DroSec_CAF1 50395 110 - 1 UCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGGGCCAACACCGCAGUUGGUCCGCACCC----CUCGAAUC------GGAUUCCACUCC ........(((((..(((.................((((((((....)))))))).......((((((((......)))))))))))..(----(.......------)))))))..... ( -31.00) >DroSim_CAF1 53213 116 - 1 UCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGGGCCAACACCACAGUUGGUCCGCACCC----CUCGAUUCCGAUUCCGAUUCCACUCC .....(((((((...(((.................((((((((....)))))))).......((((((((......)))))))))))...----.(((....))))))))))........ ( -34.20) >DroEre_CAF1 50671 90 - 1 UCCUCAACGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGAGCCACCACCACAGUUGCUCCGCACCC------------------------------ .......((((.((((..........((((.....((((((((....)))))))).......))))..........)))).)))).....------------------------------ ( -19.85) >DroYak_CAF1 55539 104 - 1 UCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGAGCCACCACCACAGUGGCUCCGCACCC----CUCGA------------UUUCACCCC .....(((((.....(((.................((((((((....)))))))).......(((((((.......))))))).)))...----.))))------------)........ ( -25.60) >DroPer_CAF1 51471 99 - 1 UCAUCGUUGAAUUAAUGCAAAUUUCGGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCA----------A-----CAGCCUAGCCGCACCCUGGCCUGUGCUC------UGAGUCCCAACC .....((((((((......))))..((((((....((((((((....))))))))(((----------.-----(((.((((.......)))).))))))..------)))))).)))). ( -25.60) >consensus UCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGGGCCAACACCACAGUUGGUCCGCACCC____CUCGA_UC_______GAUUCCACUCC .......((((.((((..........((((.....((((((((....)))))))).......))))..........)))).))))................................... (-14.06 = -15.42 + 1.36)

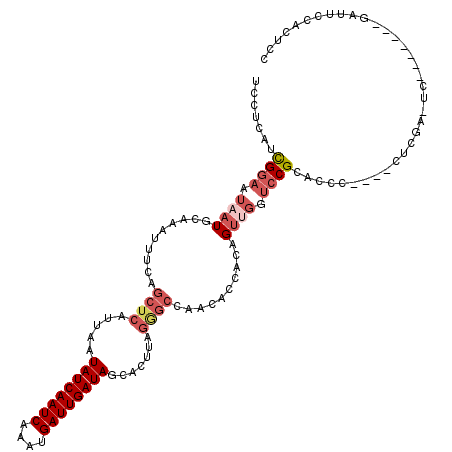
| Location | 13,424,801 – 13,424,893 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 96.96 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.54 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13424801 92 - 27905053 UAAAACAUGUGAUCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGGGCCAACACCACAGUUG ...(((.((((.(((.....)))...............((((.....((((((((....)))))))).......))))......))))))). ( -19.40) >DroSec_CAF1 50425 92 - 1 UAAAACAUGUGAUCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGGGCCAACACCGCAGUUG ...(((.((((.(((.....)))...............((((.....((((((((....)))))))).......))))......))))))). ( -19.00) >DroSim_CAF1 53249 92 - 1 UAAAACAUGUGAUCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGGGCCAACACCACAGUUG ...(((.((((.(((.....)))...............((((.....((((((((....)))))))).......))))......))))))). ( -19.40) >DroEre_CAF1 50681 92 - 1 UAAAACAUGUGAUCCUCAACGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGAGCCACCACCACAGUUG ...(((.((((.(((.....)))...............((((.....((((((((....)))))))).......))))......))))))). ( -20.30) >DroYak_CAF1 55563 91 - 1 UA-AACAUGUGAUCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGAGCCACCACCACAGUGG ..-..((((((.(((.....)))...............((((.....((((((((....)))))))).......))))......)))).)). ( -19.60) >consensus UAAAACAUGUGAUCCUCAUCGGAAUAAUGCAAAUUUCAGCUCAUUAAUAUCAAUCAAAUGAUUGAUAGCACUUAGGGCCAACACCACAGUUG ...(((.((((.(((.....)))...............((((.....((((((((....)))))))).......))))......))))))). (-19.50 = -19.30 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:58 2006