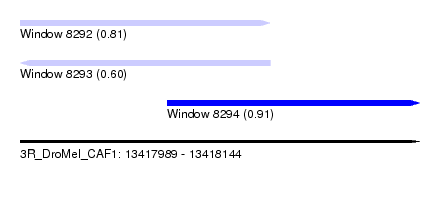
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,417,989 – 13,418,144 |
| Length | 155 |
| Max. P | 0.913506 |

| Location | 13,417,989 – 13,418,086 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -11.11 |
| Energy contribution | -11.17 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
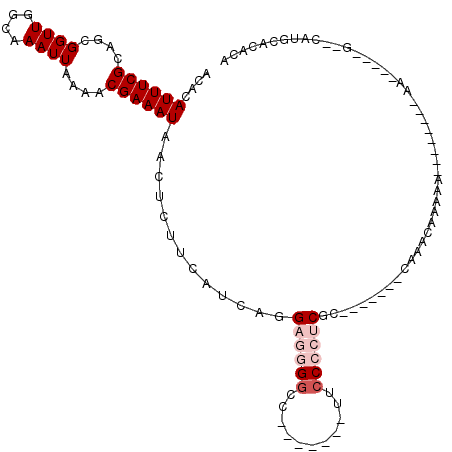
>3R_DroMel_CAF1 13417989 97 + 27905053 UUGCCUUACCACCUGCACAU-UACAACAAACAAUACUACUUUAUUGUU------GUACU-------UUUUUACACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACU (((((..(((..((((....-.......(((((((......)))))))------(((..-------....))).......)))).))).)))))................. ( -20.10) >DroSec_CAF1 43534 102 + 1 UUGCCUUACCACCUGCACAU-UGCAACAAACAAUGCUAUAUUAUUGUU------GUACUUCU--UUUUUUUACACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACU (((((..(((..((((.(((-((.......))))).........(((.------(((.....--......))))))....)))).))).)))))................. ( -18.60) >DroSim_CAF1 46041 102 + 1 UUGCCUUACCACCUGCACAU-UACAACAAACAAUGCUAUAUUAUUGUU------GUACUUCU--UUUUUUUAAACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACU (((((..(((..((((....-((((((((..............)))))------))).....--(((....)))......)))).))).)))))................. ( -18.84) >DroEre_CAF1 44213 91 + 1 UUGCCUUGCCACCUGCGCAU-UACAGCAAACAGUGCUAUUU-----UU------GUACU--------UUUUACACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACU (((((..(((..((((((..-....))(((.(((((.....-----..------)))))--------.))).........)))).))).)))))................. ( -22.60) >DroYak_CAF1 48256 96 + 1 UUGCCUUACCACCUGCGCAU-UACAGCAAACAGUGCUAUUUUAUUCUU------GUACU--------UUUUAAACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACU (((((..(((..((((((..-....))(((.(((((............------)))))--------.))).........)))).))).)))))................. ( -21.50) >DroAna_CAF1 40360 108 + 1 UUGCCGAACCACCUGCAUAUGUACA---UAUAUAUGUACAUAAGAAUUACAAAGGGGCUUGUGGGUUUUUGGAACAUUUCGCAGCGGCUGGCAAAUUAAAAUGCAAUAACU ((((((..((..((((.((((((((---(....))))))))).(((......(((((((....))))))).......))))))).)).))))))................. ( -27.32) >consensus UUGCCUUACCACCUGCACAU_UACAACAAACAAUGCUAUAUUAUUGUU______GUACU_______UUUUUAAACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACU (((((..(((..((((................................................................)))).))).)))))................. (-11.11 = -11.17 + 0.06)

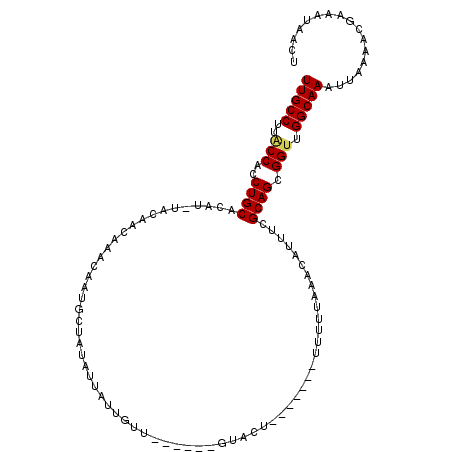

| Location | 13,417,989 – 13,418,086 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -13.79 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13417989 97 - 27905053 AGUUAUUUCGUUUUAAUUUGCCAACCGCUGCGAAAUGUGUAAAAA-------AGUAC------AACAAUAAAGUAGUAUUGUUUGUUGUA-AUGUGCAGGUGGUAAGGCAA .................(((((.(((((((((.....))).....-------.((((------(((((((((.........)))))))).-.))))).))))))..))))) ( -21.70) >DroSec_CAF1 43534 102 - 1 AGUUAUUUCGUUUUAAUUUGCCAACCGCUGCGAAAUGUGUAAAAAAA--AGAAGUAC------AACAAUAAUAUAGCAUUGUUUGUUGCA-AUGUGCAGGUGGUAAGGCAA .................(((((.((((((((....((((((......--.....)))------.)))........(((((((.....)))-))))))).)))))..))))) ( -23.80) >DroSim_CAF1 46041 102 - 1 AGUUAUUUCGUUUUAAUUUGCCAACCGCUGCGAAAUGUUUAAAAAAA--AGAAGUAC------AACAAUAAUAUAGCAUUGUUUGUUGUA-AUGUGCAGGUGGUAAGGCAA .................(((((.(((((((((...............--.....(((------(((((((((.....)))).))))))))-...)))).)))))..))))) ( -22.20) >DroEre_CAF1 44213 91 - 1 AGUUAUUUCGUUUUAAUUUGCCAACCGCUGCGAAAUGUGUAAAA--------AGUAC------AA-----AAAUAGCACUGUUUGCUGUA-AUGCGCAGGUGGCAAGGCAA .................(((((..((((((((.....((((...--------..)))------).-----..((((((.....)))))).-...)))).))))...))))) ( -23.10) >DroYak_CAF1 48256 96 - 1 AGUUAUUUCGUUUUAAUUUGCCAACCGCUGCGAAAUGUUUAAAA--------AGUAC------AAGAAUAAAAUAGCACUGUUUGCUGUA-AUGCGCAGGUGGUAAGGCAA .................(((((.(((((((((...(((......--------...))------)........((((((.....)))))).-...)))).)))))..))))) ( -22.90) >DroAna_CAF1 40360 108 - 1 AGUUAUUGCAUUUUAAUUUGCCAGCCGCUGCGAAAUGUUCCAAAAACCCACAAGCCCCUUUGUAAUUCUUAUGUACAUAUAUA---UGUACAUAUGCAGGUGGUUCGGCAA .................(((((((((((((((.................(((((....)))))......(((((((((....)---)))))))))))).)))))).))))) ( -28.70) >consensus AGUUAUUUCGUUUUAAUUUGCCAACCGCUGCGAAAUGUGUAAAAA_______AGUAC______AACAAUAAAAUAGCAUUGUUUGUUGUA_AUGUGCAGGUGGUAAGGCAA .................(((((.(((((((((...(......).............................((((((.....)))))).....)))).)))))..))))) (-13.79 = -13.85 + 0.06)

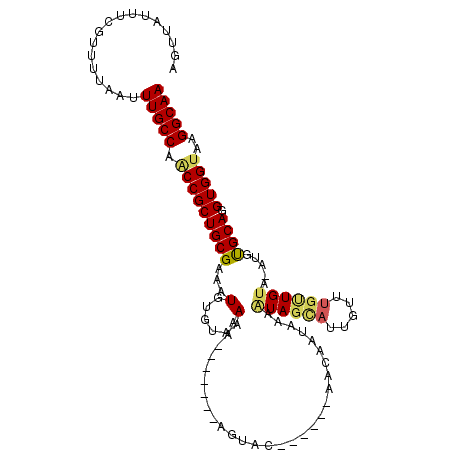
| Location | 13,418,046 – 13,418,144 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -7.20 |
| Energy contribution | -8.60 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13418046 98 + 27905053 ACACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACUCUUCAUCAGGAGGGGCC-------UUCCCCUCGC-------CAAACAAAAAAACCAAAAAA-----C--CAUGCACACA .........(((..(((((((..........(((.......))).....((((((..-------..))))))))-------)).................)-----)--).)))..... ( -22.22) >DroSec_CAF1 43596 89 + 1 ACACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACUCUUCAUCAGGAGGGGCC-------UUCCCCUCGC-------CAAACAAAA---------AA-----G--CAUGCACACA .........(((((..(((((..........(((.......))).....((((((..-------..))))))))-------)))......---------..-----)--).)))..... ( -23.20) >DroSim_CAF1 46103 89 + 1 AAACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACUCUUCAUCAGGAGGGGCC-------UUCCCCUCGC-------CAAACAAAA---------AA-----G--CAUGCACACA .........(((((..(((((..........(((.......))).....((((((..-------..))))))))-------)))......---------..-----)--).)))..... ( -23.20) >DroEre_CAF1 44264 101 + 1 ACACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACUCUUCAUCAGG---GGCC-------GUCCCCUCGCACAAAAGCAAACAAAAA--------AAUAUAUACAUAUGCAUAUA .........((.((..(((((..........(((.......)))...(((---((..-------..))))).)).)))..)).........--------.((((....))))))..... ( -16.70) >DroYak_CAF1 48312 92 + 1 AAACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACUCUUCAUCAGG---GUCCGCGGCAGGUCCCCUCGC-----------CAAAAA--------AA-----GCAUAUACACUCA .........((.....(((((..........(((.......)))....((---(.((......)).)))...))-----------)))...--------..-----))........... ( -18.70) >consensus ACACAUUUCGCAGCGGUUGGCAAAUUAAAACGAAAUAACUCUUCAUCAGGAGGGGCC_______UUCCCCUCGC_______CAAACAAAAA________AA_____G__CAUGCACACA ....((((((....((((....))))....)))))).............((((((...........))))))............................................... ( -7.20 = -8.60 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:52 2006