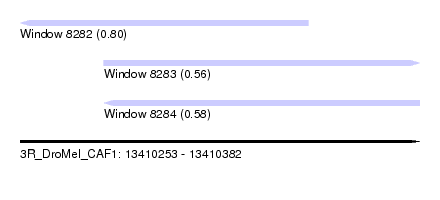
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
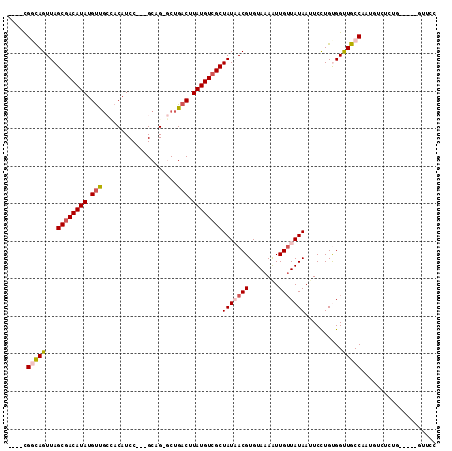
| Location | 13,410,253 – 13,410,382 |
| Length | 129 |
| Max. P | 0.796222 |

| Location | 13,410,253 – 13,410,346 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13410253 93 - 27905053 CAACUUUACACGUUAUAGCGACAUAAGUCAGC-CGGC---GGAUGUAGCAACAUAUGUUGCUAACUGCCGACUGGGGUGGCGCAUCCCGA-UCCUGAA-- ..........((..((.(((.(((...((((.-((((---((...(((((((....))))))).)))))).)))).))).)))))..)).-.......-- ( -32.50) >DroVir_CAF1 39884 94 - 1 CAAUUUUACACGUAAUAGCGACAUAAGUCAGUCCGGCAGAGGAUGUCCCGGCAUAUGUCGCUAACUGCCGCUGGGGGUGGCUCAUCCGGGAACG------ ...............((((((((((.(((.((((......)))).....))).))))))))))...(((((.....))))).....((....))------ ( -31.10) >DroGri_CAF1 39906 94 - 1 CAAUUUUACACGUAAUAGCGACAUAAGUCAG-CCAGCAAAGGAUGUCCAGACAUAUGUCGCUAACUGCCACUGG-GGUGGCUCAUUCAGCAGCCAG---- ...........((..((((((((((.(((.(-....)...((....)).))).))))))))))...(((((...-.))))).......))......---- ( -28.90) >DroEre_CAF1 36956 93 - 1 CAACUUUACACGUUAUAGCGACAUAAGUCAGC-CGGC---GGAUGUGGCAACAUAUGUCGCUAACUGCCGAUUGGGGUGGCGCAUCCUGA-UCCUGAC-- ..........(((....)))......(((((.-.(((---((..((((((.....))))))...)))))(((..(((((...)))))..)-)))))))-- ( -32.60) >DroYak_CAF1 41019 95 - 1 CAACUUUACACGUUAUAGUGACAUAAGUCAGC-CGGC---GGAUGUGGCAACAUAUGUCGCUAACUGCCGAUUGGGGUGGCGCAUCCCGA-UCCUGAUCC ........(((......)))......(((((.-.(((---((..((((((.....))))))...)))))((((((((((...))))))))-))))))).. ( -35.20) >DroAna_CAF1 33784 86 - 1 CAAUUUUACACGUUAUAGCGACAUAAGUCAGC-CUGC---GGAUGUGGCAACAUAUGUCGCUAACUACCGAUUGGGGUAGCUCAUCC--------GUC-- .........(((...((((((((((.((..((-(...---......))).)).)))))))))).(((((......)))))......)--------)).-- ( -26.20) >consensus CAACUUUACACGUUAUAGCGACAUAAGUCAGC_CGGC___GGAUGUGGCAACAUAUGUCGCUAACUGCCGAUUGGGGUGGCGCAUCCCGA_UCCUGA___ ...............((((((((((.(((.....)))......(((....))))))))))))).(((((......))))).................... (-19.48 = -19.12 + -0.36)


| Location | 13,410,280 – 13,410,382 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13410280 102 + 27905053 ----CGGCAGUUAGCAACAUAUGUUGCUACAUCC---GCCG-GCUGACUUAUGUCGCUAUAACGUGUAAAGUUGGUAUAAUUCCUGUGGUUGCCAAUGUCUCUGG----GUUCC ----.(((((.(((((((....)))))))...((---((.(-((..((((....(((......)))..))))..).......)).)))))))))...........----..... ( -27.31) >DroVir_CAF1 39908 98 + 1 ----CGGCAGUUAGCGACAUAUGCCGGGACAUCCUCUGCCGGACUGACUUAUGUCGCUAUUACGUGUAAAAUUGUUAUAAUUCUUGCGGUUGCCAUAC-----------GCUC- ----.(((((.((((((((((.(((((.(.......).)))).).....)))))))))).....((((((((((...))))).))))).)))))....-----------....- ( -27.40) >DroPse_CAF1 36342 110 + 1 UCAGUGCCAGUUAGCGACAUAUGUUGCCACAUCC---GCAG-GCUGACUUAUGUCGCUAUAACGUGUAAAAUUGUUAUAAUUCCUGUGGUUGCCAAUGUCCCUGUUCUCGUUCC .(((.(.((....(((((....)))))..((.((---((((-(..(((....)))..(((((((........)))))))...))))))).))....)).).))).......... ( -27.00) >DroGri_CAF1 39931 97 + 1 ----UGGCAGUUAGCGACAUAUGUCUGGACAUCCUUUGCUGG-CUGACUUAUGUCGCUAUUACGUGUAAAAUUGUUAUAUGUCUUGCGGUUGCCACAC-----------GUUC- ----((((((.((((((((((.(((.((.((........)).-))))).))))))))))..((((((((.....)))))))).......))))))...-----------....- ( -29.50) >DroAna_CAF1 33804 102 + 1 ----CGGUAGUUAGCGACAUAUGUUGCCACAUCC---GCAG-GCUGACUUAUGUCGCUAUAACGUGUAAAAUUGUUAUAAUUCCUGUGGCUGCCAAUGCCUCGAG----GUCCC ----.((((((((((((((((.((((((......---...)-)).))).))))))))(((((((........))))))).......))))))))...(((....)----))... ( -34.70) >DroPer_CAF1 37144 110 + 1 UCAGUGCCAGUUAGCGACAUAUGUUGCCACAUCC---GCAG-GCUGACUUAUGUCGCUAUAACGUGUAAAAUUGUUAUAAUUCCUGUGGUUGCCAAUGUCUCUGUUCUCGUUCC .(((.(.((....(((((....)))))..((.((---((((-(..(((....)))..(((((((........)))))))...))))))).))....)).).))).......... ( -27.80) >consensus ____CGGCAGUUAGCGACAUAUGUUGCCACAUCC___GCAG_GCUGACUUAUGUCGCUAUAACGUGUAAAAUUGUUAUAAUUCCUGUGGUUGCCAAUGUCUCUG_____GUUCC .....(((((...((((((((.(((....................))).))))))))(((((((........)))))))..........))))).................... (-18.10 = -18.77 + 0.67)
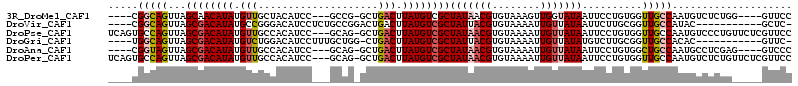
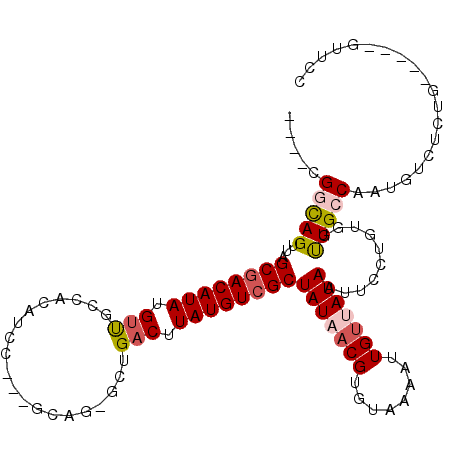
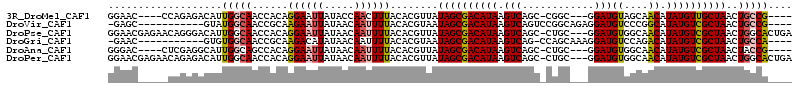
| Location | 13,410,280 – 13,410,382 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -15.11 |
| Energy contribution | -15.37 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13410280 102 - 27905053 GGAAC----CCAGAGACAUUGGCAACCACAGGAAUUAUACCAACUUUACACGUUAUAGCGACAUAAGUCAGC-CGGC---GGAUGUAGCAACAUAUGUUGCUAACUGCCG---- .....----...........(((.......((.......)).((((....(((....)))....))))..))-)(((---((...(((((((....))))))).))))).---- ( -26.90) >DroVir_CAF1 39908 98 - 1 -GAGC-----------GUAUGGCAACCGCAAGAAUUAUAACAAUUUUACACGUAAUAGCGACAUAAGUCAGUCCGGCAGAGGAUGUCCCGGCAUAUGUCGCUAACUGCCG---- -....-----------....((((..(....).......................((((((((((.(((.((((......)))).....))).))))))))))..)))).---- ( -29.40) >DroPse_CAF1 36342 110 - 1 GGAACGAGAACAGGGACAUUGGCAACCACAGGAAUUAUAACAAUUUUACACGUUAUAGCGACAUAAGUCAGC-CUGC---GGAUGUGGCAACAUAUGUCGCUAACUGGCACUGA ..........(((...(((((((..((.((((..(((((((..........))))))).(((....)))..)-))).---))(((((....)))))...))))).))...))). ( -30.40) >DroGri_CAF1 39931 97 - 1 -GAAC-----------GUGUGGCAACCGCAAGACAUAUAACAAUUUUACACGUAAUAGCGACAUAAGUCAG-CCAGCAAAGGAUGUCCAGACAUAUGUCGCUAACUGCCA---- -..((-----------(((((....)(....)...............))))))..((((((((((.(((.(-....)...((....)).))).)))))))))).......---- ( -28.60) >DroAna_CAF1 33804 102 - 1 GGGAC----CUCGAGGCAUUGGCAGCCACAGGAAUUAUAACAAUUUUACACGUUAUAGCGACAUAAGUCAGC-CUGC---GGAUGUGGCAACAUAUGUCGCUAACUACCG---- ((..(----((.(.(((.......))).)))).......................((((((((((.((..((-(...---......))).)).))))))))))....)).---- ( -28.80) >DroPer_CAF1 37144 110 - 1 GGAACGAGAACAGAGACAUUGGCAACCACAGGAAUUAUAACAAUUUUACACGUUAUAGCGACAUAAGUCAGC-CUGC---GGAUGUGGCAACAUAUGUCGCUAACUGGCACUGA ..........(((.(.(((((((..((.((((..(((((((..........))))))).(((....)))..)-))).---))(((((....)))))...))))).)).).))). ( -29.40) >consensus GGAAC_____CAGAGACAUUGGCAACCACAGGAAUUAUAACAAUUUUACACGUUAUAGCGACAUAAGUCAGC_CUGC___GGAUGUGGCAACAUAUGUCGCUAACUGCCA____ ...................(((((......((((((.....))))))........((((((((((.(((............)))((....)).))))))))))..))))).... (-15.11 = -15.37 + 0.25)
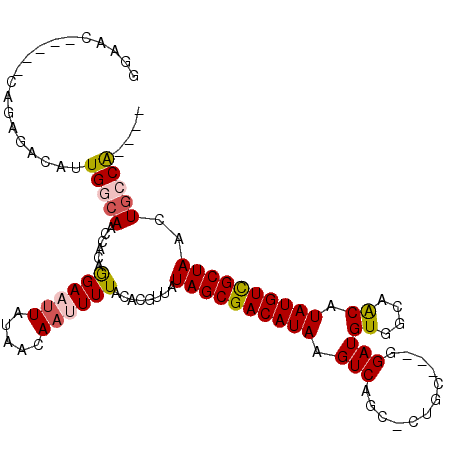
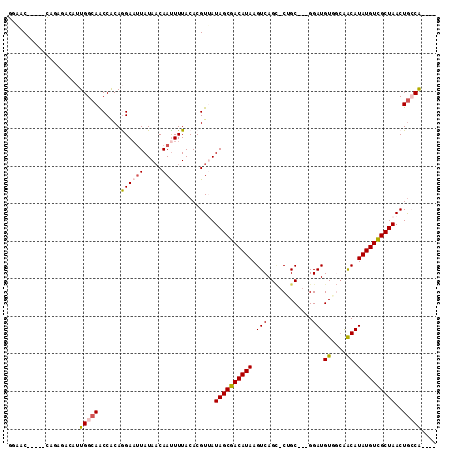
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:43 2006