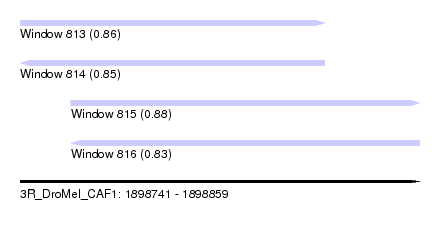
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
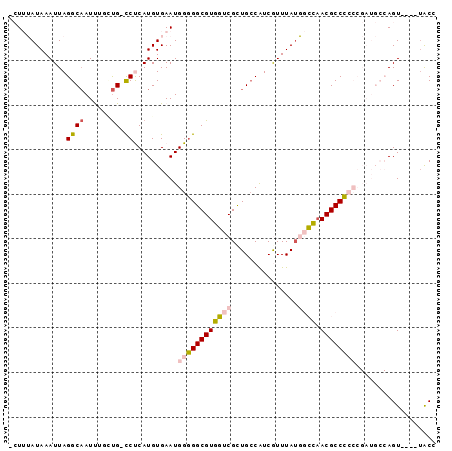
| Location | 1,898,741 – 1,898,859 |
| Length | 118 |
| Max. P | 0.880889 |

| Location | 1,898,741 – 1,898,831 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -17.00 |
| Energy contribution | -16.69 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1898741 90 + 27905053 GGCA----ACUGGCAUCGGGAGGCGUUGGCCAUAAACGAUGGCAACGACCACGCCUCCAUUCACAUGGGG-CAAAAAAUUGCCUAAUUUAUAAAG- ((((----((((....)))..(((((((.((((.....))))))))).))..(((.((((....))))))-)......)))))............- ( -29.80) >DroPse_CAF1 44412 87 + 1 CGUACGAUACU---------GGGCGUUGCUAAUAAAUGAUGGCAGCGGGCACGCCCCCAUUCACAUGAGGACACGAGAUUGCCUAAUUUAUUUAUG ((((.((((.(---------(((((((((((........)))))))(((....)))........................)))))...)))))))) ( -20.30) >DroSec_CAF1 57016 90 + 1 GGCA----ACUGGCAUCGGGGGGCGUUGGCCAUAAACGAUGCCAGCGACCACGCCCCCAUUCACAUGAGG-CAGCAAAUUGCCUAAUUUAUAAAG- ((((----((((((((((...(((....))).....))))))))).......(((..(((....))).))-)......)))))............- ( -32.70) >DroSim_CAF1 57839 90 + 1 GGCA----ACUGGCAUCGGGGGGCGUUGGCCAUAAACGAUGGCAGCGACCACGCCCCCAUUCACAUGAGG-CAGCAAAUUGCCUAAUUUAUAAAG- ((((----(...((....((((((((..(((((.....))))).......)))))))).(((....))).-..))...)))))............- ( -33.10) >DroEre_CAF1 59109 90 + 1 GGUC----ACUGGCAUCGGGGGGCGUGAGCCCUAAACGACGGCAGCGACCACGCCCCCAUUCACAUGGGA-CGGCAAAUGGCCUAAUUUAUAAAG- ((((----.(((.(.(((.(((((....)))))...))).).))).))))..((((((((....))))).-.)))....................- ( -40.70) >DroPer_CAF1 44806 87 + 1 CGUACGAUACU---------GGGCGUUGCUAAUAAAUGAUGGCAGCGGGCACGCCCCCAUUCACAUGAGGACAUGCGAUUGCCUAAUUUAUUUAUG ((((.((((.(---------(((((((((((........)))))))(((....)))....((.((((....)))).))..)))))...)))))))) ( -23.80) >consensus GGCA____ACUGGCAUCGGGGGGCGUUGGCCAUAAACGAUGGCAGCGACCACGCCCCCAUUCACAUGAGG_CAGCAAAUUGCCUAAUUUAUAAAG_ ((((..............((((((((.((...................)))))))))).(((....)))..........))))............. (-17.00 = -16.69 + -0.30)

| Location | 1,898,741 – 1,898,831 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.19 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1898741 90 - 27905053 -CUUUAUAAAUUAGGCAAUUUUUUG-CCCCAUGUGAAUGGAGGCGUGGUCGUUGCCAUCGUUUAUGGCCAACGCCUCCCGAUGCCAGU----UGCC -............(((((((.((..-(.....)..)).((((((((((((((.((....))..)))))).)))))))).......)))----)))) ( -30.90) >DroPse_CAF1 44412 87 - 1 CAUAAAUAAAUUAGGCAAUCUCGUGUCCUCAUGUGAAUGGGGGCGUGCCCGCUGCCAUCAUUUAUUAGCAACGCCC---------AGUAUCGUACG ....(((((((..((((....(((((((((((....))))))))))).....))))...)))))))..........---------........... ( -20.30) >DroSec_CAF1 57016 90 - 1 -CUUUAUAAAUUAGGCAAUUUGCUG-CCUCAUGUGAAUGGGGGCGUGGUCGCUGGCAUCGUUUAUGGCCAACGCCCCCCGAUGCCAGU----UGCC -............(((((((.((..-..((....))..(((((((((((((..(((...)))..))))).))))))))....)).)))----)))) ( -34.40) >DroSim_CAF1 57839 90 - 1 -CUUUAUAAAUUAGGCAAUUUGCUG-CCUCAUGUGAAUGGGGGCGUGGUCGCUGCCAUCGUUUAUGGCCAACGCCCCCCGAUGCCAGU----UGCC -............(((((((.((..-..((....))..(((((((((((((..((....))...))))).))))))))....)).)))----)))) ( -33.60) >DroEre_CAF1 59109 90 - 1 -CUUUAUAAAUUAGGCCAUUUGCCG-UCCCAUGUGAAUGGGGGCGUGGUCGCUGCCGUCGUUUAGGGCUCACGCCCCCCGAUGCCAGU----GACC -............(((.....)))(-((((((....)).)))))..((((((((.(((((....((((....))))..))))).))))----)))) ( -37.60) >DroPer_CAF1 44806 87 - 1 CAUAAAUAAAUUAGGCAAUCGCAUGUCCUCAUGUGAAUGGGGGCGUGCCCGCUGCCAUCAUUUAUUAGCAACGCCC---------AGUAUCGUACG ....(((((((..((((...((((((((((((....))))))))))))....))))...)))))))..........---------........... ( -27.30) >consensus _CUUUAUAAAUUAGGCAAUUUGCUG_CCUCAUGUGAAUGGGGGCGUGGUCGCUGCCAUCGUUUAUGGCCAACGCCCCCCGAUGCCAGU____UACC .............((((......)).))..........((((((((((((...............)))).)))))))).................. (-16.69 = -17.19 + 0.50)

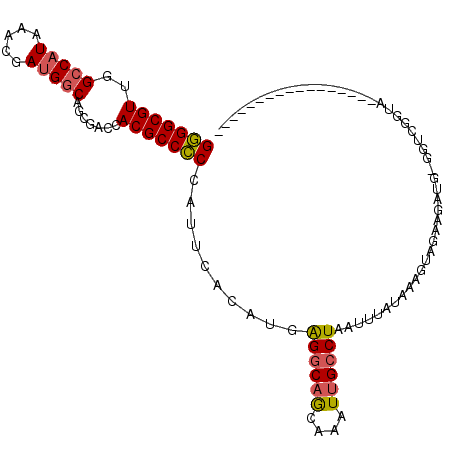
| Location | 1,898,756 – 1,898,859 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -20.01 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1898756 103 + 27905053 GAGGCGUUGGCCAUAAACGAUGGCAACGACCACGCCUCCAUUCACAUGGGGCAAAAAAUUGCCUAAUUUAUAAAGUAG-------GGUCGGUAGAUUUAACAUUAAACAU (((((((..(((((.....))))).......))))))).......(((((((((....))))))..........((..-------((((....))))..))......))) ( -28.30) >DroSec_CAF1 57031 92 + 1 GGGGCGUUGGCCAUAAACGAUGCCAGCGACCACGCCCCCAUUCACAUGAGGCAGCAAAUUGCCUAAUUUAUAAAGUAGAAGAUG-GGUCGGUA----------------- ..(((((((((.((.....))))))))).))..(((((((((......((((((....))))))..(((((...))))).))))-))..))).----------------- ( -28.80) >DroSim_CAF1 57854 92 + 1 GGGGCGUUGGCCAUAAACGAUGGCAGCGACCACGCCCCCAUUCACAUGAGGCAGCAAAUUGCCUAAUUUAUAAAGUAGAAGACG-GCUCGGUA----------------- (((((((..(((((.....))))).......)))))))..........((((((....))))))....(((..(((........-)))..)))----------------- ( -26.40) >DroEre_CAF1 59124 107 + 1 GGGGCGUGAGCCCUAAACGACGGCAGCGACCACGCCCCCAUUCACAUGGGACGGCAAAUGGCCUAAUUUAUAAAGUGGCAGAUAUGGUCGGUACCG-UAGC--GAAACAU (((((....)))))...((((((.(.((((((.((((((((....)))))..))).....(((.............))).....)))))).).)))-)..)--)...... ( -42.32) >consensus GGGGCGUUGGCCAUAAACGAUGGCAGCGACCACGCCCCCAUUCACAUGAGGCAGCAAAUUGCCUAAUUUAUAAAGUAGAAGAUG_GGUCGGUA_________________ (((((((..(((((.....))))).......)))))))..........((((((....)))))).............................................. (-20.01 = -20.20 + 0.19)

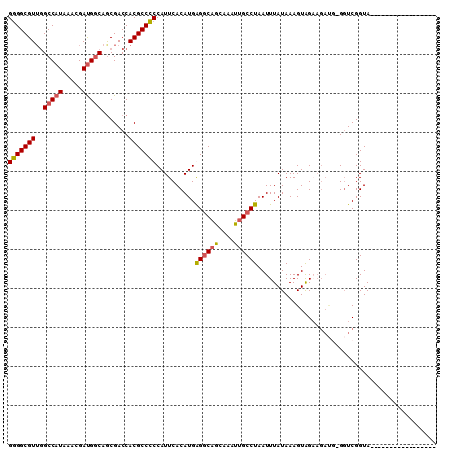
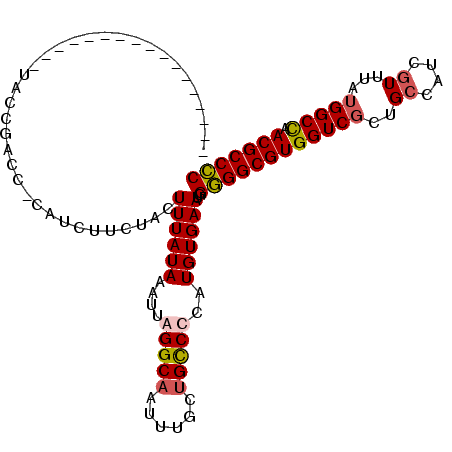
| Location | 1,898,756 – 1,898,859 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -21.73 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1898756 103 - 27905053 AUGUUUAAUGUUAAAUCUACCGACC-------CUACUUUAUAAAUUAGGCAAUUUUUUGCCCCAUGUGAAUGGAGGCGUGGUCGUUGCCAUCGUUUAUGGCCAACGCCUC .........................-------...............(((((....)))))((((....))))((((((((((((.((....))..)))))).)))))). ( -26.80) >DroSec_CAF1 57031 92 - 1 -----------------UACCGACC-CAUCUUCUACUUUAUAAAUUAGGCAAUUUGCUGCCUCAUGUGAAUGGGGGCGUGGUCGCUGGCAUCGUUUAUGGCCAACGCCCC -----------------........-..........((((((....(((((......)))))..))))))..((((((((((((..(((...)))..))))).))))))) ( -27.90) >DroSim_CAF1 57854 92 - 1 -----------------UACCGAGC-CGUCUUCUACUUUAUAAAUUAGGCAAUUUGCUGCCUCAUGUGAAUGGGGGCGUGGUCGCUGCCAUCGUUUAUGGCCAACGCCCC -----------------........-..........((((((....(((((......)))))..))))))..((((((((((((..((....))...))))).))))))) ( -27.10) >DroEre_CAF1 59124 107 - 1 AUGUUUC--GCUA-CGGUACCGACCAUAUCUGCCACUUUAUAAAUUAGGCCAUUUGCCGUCCCAUGUGAAUGGGGGCGUGGUCGCUGCCGUCGUUUAGGGCUCACGCCCC .......--((.(-(((((.(((((((....(((.............(((.....)))..(((((....))))))))))))))).)))))).))...((((....)))). ( -42.70) >consensus _________________UACCGACC_CAUCUUCUACUUUAUAAAUUAGGCAAUUUGCUGCCCCAUGUGAAUGGGGGCGUGGUCGCUGCCAUCGUUUAUGGCCAACGCCCC ....................................((((((....(((((......)))))..))))))..((((((((((((..((....))...))))).))))))) (-21.73 = -22.22 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:52 2006