
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,385,286 – 13,385,493 |
| Length | 207 |
| Max. P | 0.543510 |

| Location | 13,385,286 – 13,385,393 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -43.71 |
| Consensus MFE | -28.38 |
| Energy contribution | -27.75 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
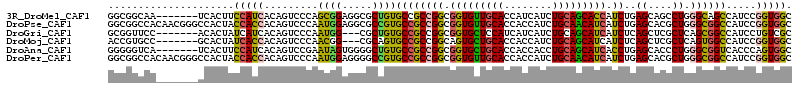
>3R_DroMel_CAF1 13385286 107 + 27905053 GGCGGCAA-------UCACUUCCAUCACAGUCCCAGCGGAGGCGCUGUGCCGCCGGCGGUGUUGCACCAUCAUCUGCAGCACCAUCUGAGCAGCCUGGGCAGCCAUCCGGUGGC (((((((.-------.(.(((((..............))))).)...)))))))((((((((((((........))))))))).........((....)).))).......... ( -44.64) >DroPse_CAF1 19756 114 + 1 GGCGGCCACAACGGGCCACUACCACCACAGUCCCAAUGGAGGCGCCGUGCCGCCGGCGGUGUUGCACCACCAUCUGCAACAUCAUCUGAGCACGCUGGGCGGCCAUCCGGUGGC (((((((......))))............(((((...)).))))))..((((((((.(((((((((........)))))))))...((.((.(((...))))))).)))))))) ( -49.40) >DroGri_CAF1 20425 104 + 1 GCGGUUCC-------ACACUAUCAUCACAGUCCCAAUGG---CGCUGUGCCGCCGGCGGUGCUCCAUCAUCAUCUGCAGCAUCAUCUCAGCUCGCUCAGCGGCCAUCCUGUCGC ((((....-------..........((((((.((...))---.))))))))))((((((((((.((........)).))))))......((.(((...)))))......)))). ( -28.53) >DroMoj_CAF1 20007 104 + 1 ACCGUGCC-------GCACUAUCACCACAGUCCCAACGG---CGCAGUGCCGCCGGCAGUGCUGCACCACCAUCUGCAGCAUCAUCUCAGCUCGCUCAGUGGCCAUCCGGUGGC ...(((((-------(.(((........))).....)))---)))...((((((((..((((((((........)))))))).......((.(((...)))))...)))))))) ( -38.00) >DroAna_CAF1 17868 107 + 1 GGGGGUCA-------UCACUUCCAUCACAGUCCGAAUAGUGGGGCUGUGCCGCCGGCGGUGCUGCACCACCACCUGCAGCAUCACCUGAGCACCCUGGGCGGUCACCCAGUGGC ((((((..-------..))))))..((((((((........))))))))..(((((.(((((((((........))))))))).)).).)).(((((((......))))).)). ( -48.20) >DroPer_CAF1 20373 114 + 1 GGCGGCCACAACGGGCCACUACCACCACAGUCCCAAUGGAGGGGCCGUGCCGCCGGCGGUGUUGCACCACCAUCUGCAACAUCAUCUGAGCACGCUGGGCGGCCAUCCGGUGGC ((.((((......)))).)).(((((((.(((((......))))).))((((((((((((((((((........)))))))))....(....)))).)))))).....))))). ( -53.50) >consensus GGCGGCCA_______CCACUACCACCACAGUCCCAAUGGAGGCGCUGUGCCGCCGGCGGUGCUGCACCACCAUCUGCAGCAUCAUCUGAGCACGCUGGGCGGCCAUCCGGUGGC .....................(((((.........((((.....))))((((((((.(((((((((........))))))))).))..((....)).)))))).....))))). (-28.38 = -27.75 + -0.63)


| Location | 13,385,393 – 13,385,493 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -39.37 |
| Consensus MFE | -23.45 |
| Energy contribution | -24.95 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13385393 100 - 27905053 CUGCUGGCCGGUGGGCGUGAGUGCGUGCUGCAGCAGGGCGGGAUGUGUGGCCCCGUGGGCGGGAUGCAAGUGGCCAAAGGUGUGGAGCGCGUGAUGCAGC ((((((((((.(..((....))((((.((((.((.((((.(......).)))).))..)))).)))).).))))))...(((.....))).....)))). ( -42.30) >DroVir_CAF1 20089 91 - 1 ---------UGUCGGCGCCACAGCAUGCUGCAACAGGGCGGGAUGCUGUGCCGCAUGGGCCGGAUGCAGAUGGCCGAAACUGUGCAGCGCAUGAUGCAGG ---------.((((((((((((((((.((((......)))).))))))))..((((((((((........))))).....))))).)))).))))..... ( -40.00) >DroPse_CAF1 19870 100 - 1 GUGGUGGGCUGUGGGGGUCAGGGCGUGCUGCAGCAGUGCGGGAUGUGUGGCCGGAUGUCCGGAGUGCAGAUGGCCGAAACUGUGCAGCGCGUGAUGCAGG ........((((....(((...((((((((((.((((((.(..((..(..((((....)))).)..))..).))....))))))))))))))))))))). ( -40.30) >DroGri_CAF1 20529 91 - 1 ---------UGUGGGCGCCACAGCAUGUUGCAGCAGCGCGGGAUGCUGUGCCGCAUGAGCCGGAUGCAGAUGUCCAAAACUGUGUAGGGCAUGAUGCAGC ---------....(((...(((((((.((((......)))).))))))))))((((.((((..((((((.(......).))))))..))).).))))... ( -35.50) >DroMoj_CAF1 20111 91 - 1 ---------UGUCGGCGCAAUGGCGUGGUGCAGCAGAGCGGGAUGCGAUGCGGUGUGAGCCGGAUGCAGAUGGCCAAAGCUGUGCAGCGCGUGGUGCAGC ---------.....((((....((((.(..((((...((.(..(((.((.((((....)))).)))))..).))....))))..).))))...))))... ( -37.80) >DroPer_CAF1 20487 100 - 1 GUGGUGGGCUGUGGGGGUCAGGGCGUGCUGCAGCAGUGCGGGAUGUGUGGCCGGAUGUCCGGAGUGCAGAUGGCCGAAACUGUGCAGCGCGUGAUGCAGG ........((((....(((...((((((((((.((((((.(..((..(..((((....)))).)..))..).))....))))))))))))))))))))). ( -40.30) >consensus _________UGUGGGCGCCAGGGCGUGCUGCAGCAGGGCGGGAUGCGUGGCCGCAUGAGCCGGAUGCAGAUGGCCAAAACUGUGCAGCGCGUGAUGCAGC ..............(((((...((((((((((.(((....((.(((....((((....))))...))).....))....))))))))))))))))))... (-23.45 = -24.95 + 1.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:34 2006