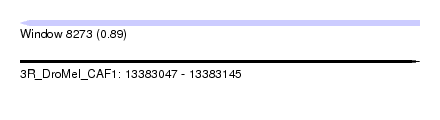
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,383,047 – 13,383,145 |
| Length | 98 |
| Max. P | 0.894591 |

| Location | 13,383,047 – 13,383,145 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
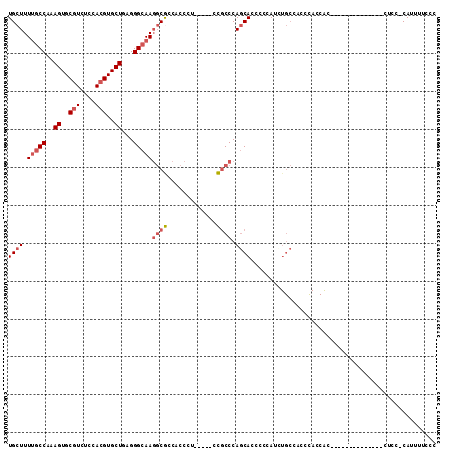
| Mean pairwise identity | 79.02 |
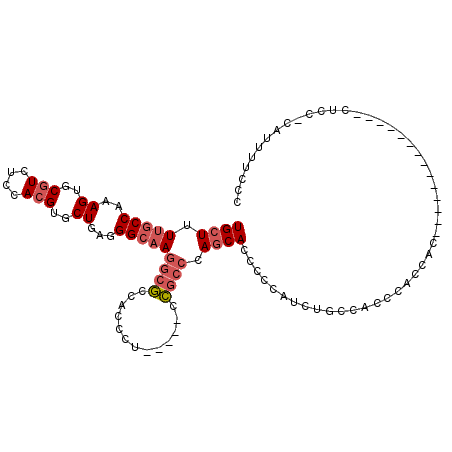
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -16.43 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
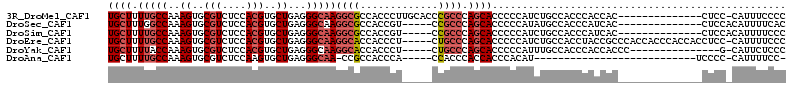
>3R_DroMel_CAF1 13383047 98 - 27905053 UGCUUUUGCCAAAGUGCGUCUCCACGUGCUGAGGGCAAGGCGCCACCCUUGCACCCGCCCAGCACCCCCAUCUGCCACCCACCAC--------------CUCC-CAUUUCCCC .((....))...((..(((....)))..))((((((((((......)))))).........(((........))).........)--------------))).-......... ( -22.90) >DroSec_CAF1 16516 94 - 1 UGCUUUGGCCAAAGUGCGUCUCCACGUGCUGAGGGCAAGGCGCCACCGU-----CCGCCCAGCACCCCCAUAUGCCACCCAUCAC--------------CUCCACAUUUUCAC .....((((....(((......)))(((((..((((..((((....)))-----).)))))))))........))))........--------------.............. ( -23.50) >DroSim_CAF1 17190 94 - 1 UGCUUUUGCCAAAGUGCGUCUCCACGUGCUGAGGGCAAGGCGCCACCGU-----CCGCCCAGCACCCCCAUCUGCCACCCAUCAC--------------CUCCACAUUUUCCC ((((.(((((..((..(((....)))..))...)))))((((.......-----.)))).)))).....................--------------.............. ( -23.60) >DroEre_CAF1 17141 107 - 1 UGCUUUUGCCAAAGUGCGUCUCCACGUGCUGAGGGCAAGGCACCACCCU-----CUGCCCAGCACCCCCAUCUGCCACCUACCGCCCACCACCCACCACCUCC-CAUUUUCCC ((((.(((((..((..(((....)))..))...)))))((((.......-----.)))).)))).......................................-......... ( -22.80) >DroYak_CAF1 21024 92 - 1 UGCUUUUACCAAAGUGCGUCUCCACGUGCUGAGGGCAAGGCACCACCCU-----CUGCCCAGCACCCCCAUUUGCCACCCACCACCC---------------G-CAUUCUCCC (((.........((..(((....)))..))..((((((((......)))-----.))))).(((........)))............---------------)-))....... ( -21.60) >DroAna_CAF1 15699 78 - 1 UGCUUUUGCCAAAGUGCGUCUCCAAGUGCUGAGGGCAA-CCGCCACCCA-----CCACCCACCACCCACAU---------------------------UCCCC-CAUUUUCC- .((..(((((..((..(........)..))...)))))-..))......-----.................---------------------------.....-........- ( -14.90) >consensus UGCUUUUGCCAAAGUGCGUCUCCACGUGCUGAGGGCAAGGCGCCACCCU_____CCGCCCAGCACCCCCAUCUGCCACCCACCAC______________CUCC_CAUUUUCCC ((((.(((((..((..(((....)))..))...)))))((((.............)))).))))................................................. (-16.43 = -17.15 + 0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:32 2006