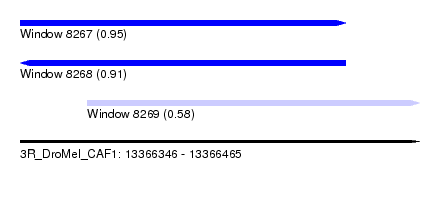
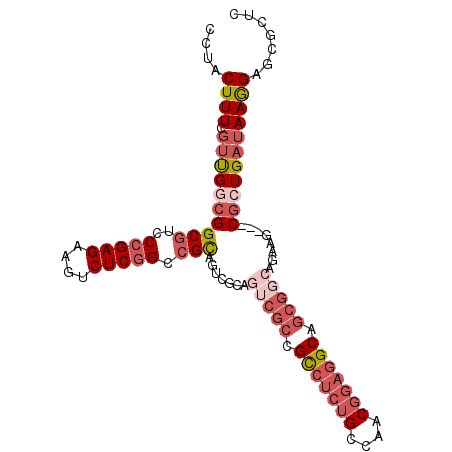
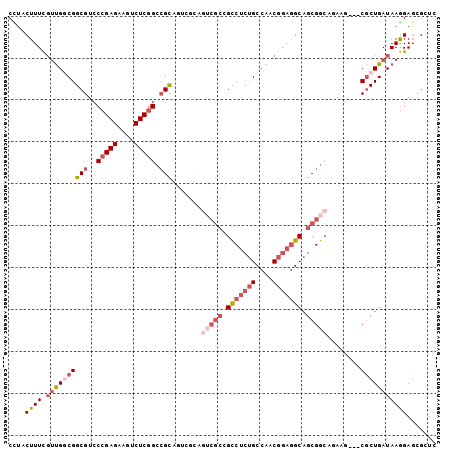
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,366,346 – 13,366,465 |
| Length | 119 |
| Max. P | 0.951127 |

| Location | 13,366,346 – 13,366,443 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -47.05 |
| Consensus MFE | -34.65 |
| Energy contribution | -37.10 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
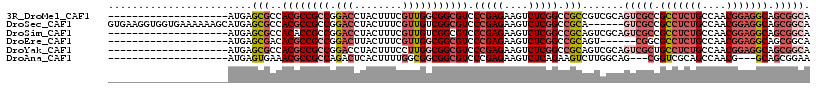
>3R_DroMel_CAF1 13366346 97 + 27905053 --------------------AUGAGCGCCACGCCGCCGGACCUACUUUCGUUGGCGGCGUCCCGAGAAGUCUCGGCCGCCGUCGCAGUCGCCGCCUCUGCCAACGGAGGCAGCGGCA --------------------....(((.(..((((.((((......)))).))))((((..(((((....))))).))))).))).(((((.(((((((....))))))).))))). ( -51.10) >DroSec_CAF1 45 111 + 1 GUGAAGGUGGUGAAAAAAGCAUGAGCGCCACGCCGCCGGACCUACUUUCGUUGUCGGCGUCCCGAGAAGUCUCGGCCGCA------GUCGCCGCCUCUGCCAACGGAGGCAGCGGCA .....(((((((......((....))....))))))).(((..((....)).))).(((..(((((....))))).))).------(((((.(((((((....))))))).))))). ( -49.20) >DroSim_CAF1 1 97 + 1 --------------------AUGAGCGCCACACCGCCGGACCUACUUUCGUUGUCGGCGUCCCGAGAAGUCUCGGCCGCAGUCGCAGUCGCCGCCUCUGCCAACGGAGGCAGCGGCA --------------------....(((.(....((((((....((....))..))))))..(((((....))))).....).))).(((((.(((((((....))))))).))))). ( -42.80) >DroEre_CAF1 1 91 + 1 --------------------AUGAGCGACACGCCGCCGGACUUACUUUCGUUGGCGGCGUCCCGAGAAGUCUCGGCCGCAGU------CGGCGCCUCUGCCAACGGAGGCAGCGGCA --------------------....(((..(((((((((.((........))))))))))).(((((....))))).))).((------((..(((((((....)))))))..)))). ( -46.20) >DroYak_CAF1 1 97 + 1 --------------------AUGAGCGCCACGCCGCCGGACCUACUUUCCUUGGCGGCGUCCCGAGAAGUCUCGGCCGCAGUCGCAGUCGCUGCCUCUGCCAACGGAGGCAGCGGCA --------------------....(((.((((((((((((.......)))..)))))))).(((((....))))).....).))).(((((((((((((....))))))))))))). ( -55.60) >DroAna_CAF1 1 91 + 1 --------------------AUGAGUGAAACGCCGCCAGACUCACUUUUGGCGGCGGCGUCCCGAGAAGUCUCAGAAGUCUUGGCAG---CGGUCGCAGCCAACG---GCAGCGGAA --------------------..........((((((((((......))))))))))...(((((((....)))....((((((((.(---(....)).))))).)---)).).))). ( -37.40) >consensus ____________________AUGAGCGCCACGCCGCCGGACCUACUUUCGUUGGCGGCGUCCCGAGAAGUCUCGGCCGCAGUCGCAGUCGCCGCCUCUGCCAACGGAGGCAGCGGCA ........................(((..((((((((.(((........))))))))))).(((((....))))).))).......(((((.(((((((....))))))).))))). (-34.65 = -37.10 + 2.45)
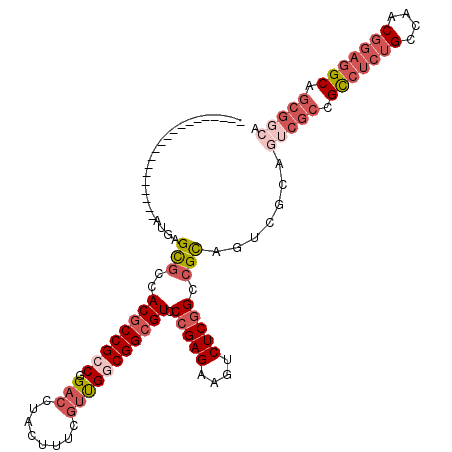
| Location | 13,366,346 – 13,366,443 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -47.08 |
| Consensus MFE | -34.60 |
| Energy contribution | -35.88 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13366346 97 - 27905053 UGCCGCUGCCUCCGUUGGCAGAGGCGGCGACUGCGACGGCGGCCGAGACUUCUCGGGACGCCGCCAACGAAAGUAGGUCCGGCGGCGUGGCGCUCAU-------------------- .(((((.(((.(((..(((...(((((((.((((....))))(((((....)))))..))))))).((....))..)))))).))))))))......-------------------- ( -53.00) >DroSec_CAF1 45 111 - 1 UGCCGCUGCCUCCGUUGGCAGAGGCGGCGAC------UGCGGCCGAGACUUCUCGGGACGCCGACAACGAAAGUAGGUCCGGCGGCGUGGCGCUCAUGCUUUUUUCACCACCUUCAC .(((.(((((......))))).)))((((.(------..((.(((((....)))))..(((((((.((....))..)).))))).))..)))))....................... ( -48.80) >DroSim_CAF1 1 97 - 1 UGCCGCUGCCUCCGUUGGCAGAGGCGGCGACUGCGACUGCGGCCGAGACUUCUCGGGACGCCGACAACGAAAGUAGGUCCGGCGGUGUGGCGCUCAU-------------------- ...(((((((((.(....).)))))))))...(((.(..((.(((((....)))))..(((((((.((....))..)).))))).))..))))....-------------------- ( -48.10) >DroEre_CAF1 1 91 - 1 UGCCGCUGCCUCCGUUGGCAGAGGCGCCG------ACUGCGGCCGAGACUUCUCGGGACGCCGCCAACGAAAGUAAGUCCGGCGGCGUGUCGCUCAU-------------------- .(((.(((((......))))).)))((.(------((.....(((((....))))).((((((((.((....))......)))))))))))))....-------------------- ( -46.30) >DroYak_CAF1 1 97 - 1 UGCCGCUGCCUCCGUUGGCAGAGGCAGCGACUGCGACUGCGGCCGAGACUUCUCGGGACGCCGCCAAGGAAAGUAGGUCCGGCGGCGUGGCGCUCAU-------------------- .(((((((((((.(....).))))))))..((((....))))(((((....))))).((((((((..(((.......))))))))))))))......-------------------- ( -51.50) >DroAna_CAF1 1 91 - 1 UUCCGCUGC---CGUUGGCUGCGACCG---CUGCCAAGACUUCUGAGACUUCUCGGGACGCCGCCGCCAAAAGUGAGUCUGGCGGCGUUUCACUCAU-------------------- ...((((((---(..((((.((....)---).))))(((((((((((....)))))))......(((.....))).)))))))))))..........-------------------- ( -34.80) >consensus UGCCGCUGCCUCCGUUGGCAGAGGCGGCGACUGCGACUGCGGCCGAGACUUCUCGGGACGCCGCCAACGAAAGUAGGUCCGGCGGCGUGGCGCUCAU____________________ ((((.(((((......))))).))))............(((.(((((....))))).((((((((...............))))))))..)))........................ (-34.60 = -35.88 + 1.28)

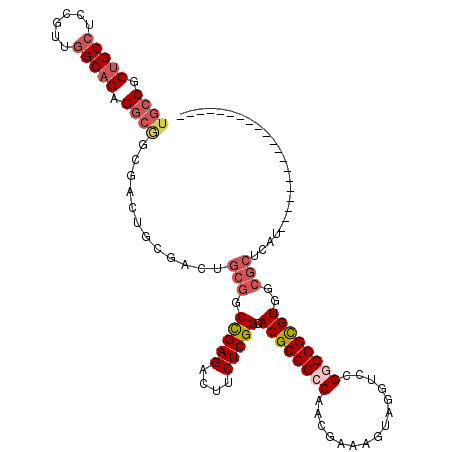
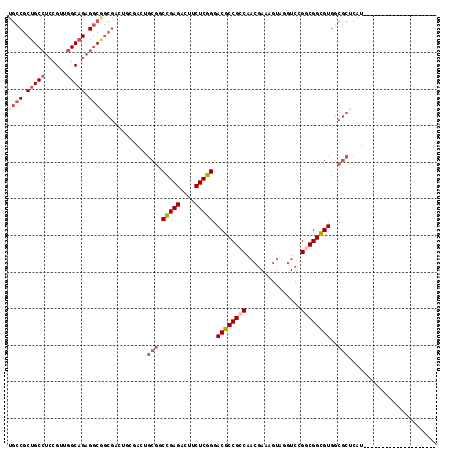
| Location | 13,366,366 – 13,366,465 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -44.98 |
| Consensus MFE | -29.09 |
| Energy contribution | -31.53 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13366366 99 + 27905053 CCUACUUUCGUUGGCGGCGUCCCGAGAAGUCUCGGCCGCCGUCGCAGUCGCCGCCUCUGCCAACGGAGGCAGCGGCAGAAG---CGCUGAUAAGGAGCGUUC ......(((((.(((((((..(((((....))))).))))))))).(((((.(((((((....))))))).))))).)))(---((((.......))))).. ( -50.10) >DroSec_CAF1 85 93 + 1 CCUACUUUCGUUGUCGGCGUCCCGAGAAGUCUCGGCCGCA------GUCGCCGCCUCUGCCAACGGAGGCAGCGGCAGAAG---CGCUGAUAAGGAGCGCUC ......(((.((((((((((.(((((....))))).....------(((((.(((((((....))))))).)))))....)---))))))))).)))..... ( -45.90) >DroSim_CAF1 21 99 + 1 CCUACUUUCGUUGUCGGCGUCCCGAGAAGUCUCGGCCGCAGUCGCAGUCGCCGCCUCUGCCAACGGAGGCAGCGGCAGAAA---CGCUGAUAAGGAGCGCUC ......(((.((((((((((.(((((....)))))..((....)).(((((.(((((((....))))))).)))))....)---))))))))).)))..... ( -46.50) >DroEre_CAF1 21 93 + 1 CUUACUUUCGUUGGCGGCGUCCCGAGAAGUCUCGGCCGCAGU------CGGCGCCUCUGCCAACGGAGGCAGCGGCAAAAG---CACUGAUAAGGAGCGCUC .........((((((.(((..(((((....))))).))).))------))))(((.(((((......))))).)))...((---(.((.......)).))). ( -39.00) >DroYak_CAF1 21 99 + 1 CCUACUUUCCUUGGCGGCGUCCCGAGAAGUCUCGGCCGCAGUCGCAGUCGCUGCCUCUGCCAACGGAGGCAGCGGCAAAAG---CGCUGAUAAGGAUCGAUC ....(..((((((.((((((.(((((....)))))..((....)).(((((((((((((....)))))))))))))....)---))))).))))))..)... ( -50.70) >DroAna_CAF1 21 96 + 1 CUCACUUUUGGCGGCGGCGUCCCGAGAAGUCUCAGAAGUCUUGGCAG---CGGUCGCAGCCAACG---GCAGCGGAAGAAGCGGCGCUGAUAAAGAGCGCUC ....(((((((((((.((...((((((..((...))..))))))..)---).))))).(((...)---))..))))))....((((((.......)))))). ( -37.70) >consensus CCUACUUUCGUUGGCGGCGUCCCGAGAAGUCUCGGCCGCAGUCGCAGUCGCCGCCUCUGCCAACGGAGGCAGCGGCAGAAG___CGCUGAUAAGGAGCGCUC ....((((.((((((((((..(((((....))))).))).......(((((.(((((((....))))))).)))))........)))))))))))....... (-29.09 = -31.53 + 2.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:29 2006