| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,360,268 – 13,360,387 |
| Length | 119 |
| Max. P | 0.895509 |

| Location | 13,360,268 – 13,360,387 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -20.35 |
| Energy contribution | -23.76 |
| Covariance contribution | 3.41 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13360268 119 + 27905053 AAGAGUUUAGUGGUACCGUGUGCGGAAGUAAGUUUCAUCUCCAGUUUAGUGUUACCAGA-GUCUAUGUUGGCUUCCUCACCACGUGGUGAAGGAGGGAAACUUUGCUGCAUACGGUGAGA .............((((((((((((....(((((((.(((((...............((-(((......)))))..(((((....))))).))))))))))))..))))))))))))... ( -45.90) >DroSec_CAF1 105295 119 + 1 AGGAAUUUAGUGGUACCGUGUGCAGAAAUUAGUUUCAUCUCCAAUUUAGUGUUACCAGA-GUCUAUGUUGUCUUGCUCACCACGUGGUGAAGGAGGGAAACUUUGCUGCAUACGGUGAAA .............((((((((((((.....((((((.(((((...............((-(.(......).)))..(((((....))))).)))))))))))...))))))))))))... ( -42.70) >DroSim_CAF1 110917 119 + 1 AGGAAUUUAGUGGUACCGUGUGCAGAAAUUAGUUUCAUCUCCAAUUUAGUGUUACCAGA-GUCUAUGUUGUCUUGCUCACCACGUGGUGAAGGAGGGAAACUUUGCUACAUACGGUAAAA .............(((((((((.((.....((((((.(((((...............((-(.(......).)))..(((((....))))).)))))))))))...)).)))))))))... ( -36.40) >DroEre_CAF1 112909 107 + 1 AAGAUGUUAGUGGUACCAUAUGCGGAAACUAGUUUCAUC-CCAAUUCAGUGUUACCAGA-GAUUAAGGUGUCUUCCUCACCACGUGGUGA-----GAAAAUUCAAUAAAAAU-----U-A ..((((..(.((((.((......))..)))).)..))))-.................((-(((......))))).((((((....)))))-----)................-----.-. ( -20.40) >DroYak_CAF1 129800 119 + 1 AAGAAGUUAGUGGUACCAUAUGUUGAAACUAGUUUUAUCCCCAAUUCAGCGUUACCUCUCUGGUAUGUUGUCUUUCUCACCACGUGGUGUAGGACGGAAACUUAGCUACAGUAGGUGA-A .........(((....))).(((((((.................)))))))((((((((.((((.....(((((...((((....)))).)))))(....)...)))).)).))))))-. ( -27.83) >consensus AAGAAUUUAGUGGUACCGUGUGCAGAAAUUAGUUUCAUCUCCAAUUUAGUGUUACCAGA_GUCUAUGUUGUCUUCCUCACCACGUGGUGAAGGAGGGAAACUUUGCUACAUACGGUGA_A .............((((((((((((.....((((((.(((((..............((....))............(((((....))))).)))))))))))...))))))))))))... (-20.35 = -23.76 + 3.41)


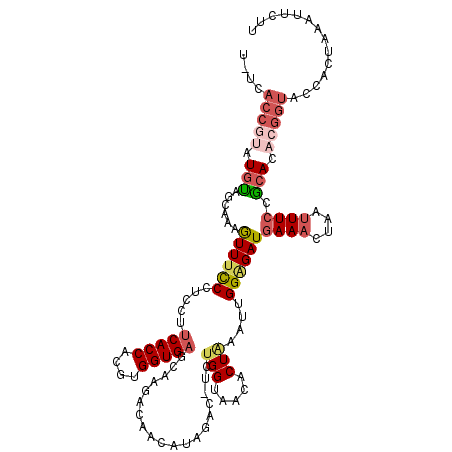
| Location | 13,360,268 – 13,360,387 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -13.36 |
| Energy contribution | -15.00 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13360268 119 - 27905053 UCUCACCGUAUGCAGCAAAGUUUCCCUCCUUCACCACGUGGUGAGGAAGCCAACAUAGAC-UCUGGUAACACUAAACUGGAGAUGAAACUUACUUCCGCACACGGUACCACUAAACUCUU ....(((((.(((.(..(((((((..((((.((((....))))))))............(-((..((........))..)))..)))))))....).))).))))).............. ( -33.90) >DroSec_CAF1 105295 119 - 1 UUUCACCGUAUGCAGCAAAGUUUCCCUCCUUCACCACGUGGUGAGCAAGACAACAUAGAC-UCUGGUAACACUAAAUUGGAGAUGAAACUAAUUUCUGCACACGGUACCACUAAAUUCCU ....(((((.(((((.((((((((.((..((((((....))))))..))..........(-((..((........))..)))..))))))..)).))))).))))).............. ( -33.50) >DroSim_CAF1 110917 119 - 1 UUUUACCGUAUGUAGCAAAGUUUCCCUCCUUCACCACGUGGUGAGCAAGACAACAUAGAC-UCUGGUAACACUAAAUUGGAGAUGAAACUAAUUUCUGCACACGGUACCACUAAAUUCCU ...((((((.(((((.((((((((.((..((((((....))))))..))..........(-((..((........))..)))..))))))..)).))))).))))))............. ( -32.50) >DroEre_CAF1 112909 107 - 1 U-A-----AUUUUUAUUGAAUUUUC-----UCACCACGUGGUGAGGAAGACACCUUAAUC-UCUGGUAACACUGAAUUGG-GAUGAAACUAGUUUCCGCAUAUGGUACCACUAACAUCUU .-.-----............(((((-----(((((....))))))))))...........-..(((((.....(((((((-.......)))))))(((....)))))))).......... ( -21.60) >DroYak_CAF1 129800 119 - 1 U-UCACCUACUGUAGCUAAGUUUCCGUCCUACACCACGUGGUGAGAAAGACAACAUACCAGAGAGGUAACGCUGAAUUGGGGAUAAAACUAGUUUCAACAUAUGGUACCACUAACUUCUU .-..(((...((((((((.((((..(((((.((((....))))............((((.....))))...........))))).)))))))))...)))...))).............. ( -23.60) >consensus U_UCACCGUAUGUAGCAAAGUUUCCCUCCUUCACCACGUGGUGAGCAAGACAACAUAGAC_UCUGGUAACACUAAAUUGGAGAUGAAACUAAUUUCCGCACACGGUACCACUAAAUUCUU ....(((((.(((......((((((.....(((((....)))))...................(((.....)))....))))))((((....)))).))).))))).............. (-13.36 = -15.00 + 1.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:25 2006