
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,354,298 – 13,354,398 |
| Length | 100 |
| Max. P | 0.858965 |

| Location | 13,354,298 – 13,354,398 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -13.41 |
| Energy contribution | -14.72 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
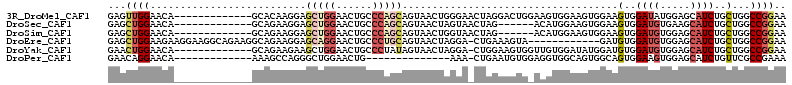
>3R_DroMel_CAF1 13354298 100 + 27905053 GAGUUGGAACA-------------GCACAAGGAGCUGGAACUGCCCAGCAGUAACUGGGAACUAGGACUGGAAGUGGAAGUGGAAGUGGAUAUGGAGCAUCUGCUGGCCGGAA .((((....((-------------((.......))))......(((((......)))))))))....((((.((..((.((....((....))...)).))..))..)))).. ( -26.20) >DroSec_CAF1 99077 94 + 1 GAGCUGGAACA-------------GCAGAAGGAGCUGGAACUGCCCAGCAGUAACUAGUAACUAG------ACAUGGAAGUGGAAGUGGAUGUGAAGCAUCUGCUGGCCGGAA ...((((..((-------------(((((....(((((......))))).((.((.....(((..------.(((....)))..)))....))...)).))))))).)))).. ( -28.40) >DroSim_CAF1 104791 94 + 1 GAGCUGGAACA-------------GCAGAAGGAGCUGGAACUGCCCAGCAGUAACUGGUAACUAG------ACAUGGAAGUGGAAGUGGAUGUGGAGCAUCUGCUGGCCGGAA ..((((...))-------------)).......(((((......))))).....(((((......------.(((....)))..((..((((.....))))..)).))))).. ( -29.30) >DroEre_CAF1 106731 100 + 1 GAGCUGGAAGAAGGAAGGCAGAAGGCAGAAGGAGCAGGAACUGCCCUGCAGUAACUAGGA-CUGAAAGUA------------GAUGUGGAUGUGGAGCAUCUGCUGGCCGGAA ...((((....((....((((..(((((............))))))))).....))....-.....((((------------(((((.........)))))))))..)))).. ( -27.50) >DroYak_CAF1 116656 99 + 1 GAACUGGAACA-------------GCAGAAGAAGCUGGAACUGCCCUAUAGUAACUAGGA-CUGGAAGUGGUUGUGGAUAUGGAUGUGGAUGUGGAGCAUCUGCUGGCCGGAA ...((((..((-------------(((((.(...(((.(.(..(.(((((....(((.((-((......)))).))).)))))..)..).).)))..).))))))).)))).. ( -30.00) >DroPer_CAF1 107787 85 + 1 GAACAGGAACA-------------AAAGCCAGGGCUGGAACUG--------------AAA-CUGAAUGUGGAGGUGGCAGUGGCAGUGGAAGUGGAGCAUCUGUUCGCCGAAA (((((((..((-------------....(((..(((...((((--------------..(-((.........)))..)))))))..)))...)).....)))))))....... ( -22.30) >consensus GAGCUGGAACA_____________GCAGAAGGAGCUGGAACUGCCCAGCAGUAACUAGGA_CUAGAAGUG_ACGUGGAAGUGGAAGUGGAUGUGGAGCAUCUGCUGGCCGGAA ...((((..........................(((((......)))))....................................(..((((.....))))..)...)))).. (-13.41 = -14.72 + 1.31)
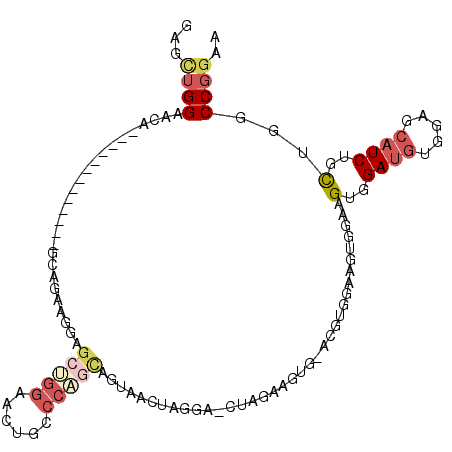
| Location | 13,354,298 – 13,354,398 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.72 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -6.45 |
| Energy contribution | -7.57 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13354298 100 - 27905053 UUCCGGCCAGCAGAUGCUCCAUAUCCACUUCCACUUCCACUUCCAGUCCUAGUUCCCAGUUACUGCUGGGCAGUUCCAGCUCCUUGUGC-------------UGUUCCAACUC ....((.(((((.(.(((..............(((...((.....))...))).((((((....)))))).......)))....).)))-------------))..))..... ( -20.20) >DroSec_CAF1 99077 94 - 1 UUCCGGCCAGCAGAUGCUUCACAUCCACUUCCACUUCCAUGU------CUAGUUACUAGUUACUGCUGGGCAGUUCCAGCUCCUUCUGC-------------UGUUCCAGCUC ....((.(((((((.(((..((((..............))))------..)))...........((((((....))))))....)))))-------------))..))..... ( -22.34) >DroSim_CAF1 104791 94 - 1 UUCCGGCCAGCAGAUGCUCCACAUCCACUUCCACUUCCAUGU------CUAGUUACCAGUUACUGCUGGGCAGUUCCAGCUCCUUCUGC-------------UGUUCCAGCUC ....((.(((((((.(((..((....(((...((......))------..)))..(((((....)))))...))...)))....)))))-------------))..))..... ( -23.00) >DroEre_CAF1 106731 100 - 1 UUCCGGCCAGCAGAUGCUCCACAUCCACAUC------------UACUUUCAG-UCCUAGUUACUGCAGGGCAGUUCCUGCUCCUUCUGCCUUCUGCCUUCCUUCUUCCAGCUC ....((...(((((.((..............------------......(((-(.......)))).(((((((...)))).)))...))..)))))...))............ ( -18.40) >DroYak_CAF1 116656 99 - 1 UUCCGGCCAGCAGAUGCUCCACAUCCACAUCCAUAUCCACAACCACUUCCAG-UCCUAGUUACUAUAGGGCAGUUCCAGCUUCUUCUGC-------------UGUUCCAGUUC ....((.(((((((.....................................(-(((((.......))))))(((....)))...)))))-------------))..))..... ( -20.30) >DroPer_CAF1 107787 85 - 1 UUUCGGCGAACAGAUGCUCCACUUCCACUGCCACUGCCACCUCCACAUUCAG-UUU--------------CAGUUCCAGCCCUGGCUUU-------------UGUUCCUGUUC ...(((.(((((((.(((...((...((((..((((.............)))-)..--------------))))...))....))).))-------------))))))))... ( -17.52) >consensus UUCCGGCCAGCAGAUGCUCCACAUCCACUUCCACUUCCACGU_CACUUCCAG_UCCUAGUUACUGCUGGGCAGUUCCAGCUCCUUCUGC_____________UGUUCCAGCUC ....((..((((((((.....)))).......................................((((((....))))))......................))))))..... ( -6.45 = -7.57 + 1.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:23 2006