
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,331,533 – 13,331,674 |
| Length | 141 |
| Max. P | 0.929632 |

| Location | 13,331,533 – 13,331,638 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -44.95 |
| Consensus MFE | -30.38 |
| Energy contribution | -29.22 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
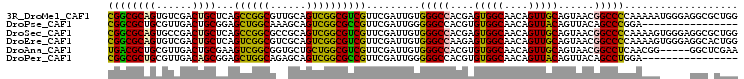
>3R_DroMel_CAF1 13331533 105 - 27905053 CGGCGCAGUGUCGACUGCUCAGCCGGCGUUGCAGUCGGCGUCGUUCGAUUGUGGGCCACGAGUGGCAACAGUUGCAGUAACGGCCCCAAAAAUGGGAGGCGCUGG ((((((...(((((((((...(....)...)))))))))(((((((((((((..((((....)))).)))))))....))))))((((....))))..)))))). ( -51.40) >DroPse_CAF1 99506 89 - 1 CGGCGCUGCGUUGACUGCGGAGCUGGCAAAGCAGUCGGCGCCGUUCGAUUGGGGGCCACGUGUGGCAACAGUUACAGUUACAGCCCGGA---------------- (((.((((...(((((((((.((((((......)))))).)))...(((((...((((....))))..))))).)))))))))))))..---------------- ( -39.50) >DroSec_CAF1 83980 105 - 1 CGGCGCAGUGCCGACUGCUCAGCCGGCGCCGCAGUCGGCGUCGUUCGAUUGUGGGCCACGAGUGGCAACAGUUGCAGUAACGGCCCCAAAAGUGGGAGGCGCUGG ((((((...(((((((((.....((((((((....))))))))...((((((..((((....)))).)))))))))))..))))((((....))))..)))))). ( -54.20) >DroEre_CAF1 90231 105 - 1 CGGCGCAGUGUCGACUGCUCAGUCGGCGUCGCAGUCGGCGUCGUUCGAUUGUGGGCCAAGAGUGGCAACAGUUGCAGUAACGGCCCCAAAAGUGGGAGGCACUGG .....(((((((..(..(((((((((((.(((.....))).)).))))))).(((((.....((....))((((...)))))))))....))..)..))))))). ( -45.20) >DroAna_CAF1 103605 100 - 1 UGACGCUGCGUUGACUGCGAAGUCGGCGGUGCUGCUGGCGUCGUUCGAUUGUGGGCCACGUGUGGCAACAGUUGCAGUAACGGCCUCAACGG-----GGCUCGAA .((.(((.((((((.......(((((((....)))))))(((((((((((((..((((....)))).)))))))....)))))).)))))).-----)))))... ( -43.10) >DroPer_CAF1 93643 89 - 1 CGGCGCUGCGUUGACAGCGGAGCUGGCAGAGCAGUCGGCGCCGUUCGAUUGGGGGCCACGUGUGGCAACAGUUACAGUUACAGCCUGGA---------------- (((.((((...(((((((((.((((((......)))))).))))).(((((...((((....))))..)))))...)))))))))))..---------------- ( -36.30) >consensus CGGCGCAGCGUCGACUGCGCAGCCGGCGGAGCAGUCGGCGUCGUUCGAUUGUGGGCCACGAGUGGCAACAGUUGCAGUAACGGCCCCAAAAG_____GGC_CUGG ((((((((......))))...((((((......)))))))))).........(((((....(((((....)))))......)))))................... (-30.38 = -29.22 + -1.16)


| Location | 13,331,560 – 13,331,674 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -47.22 |
| Consensus MFE | -31.69 |
| Energy contribution | -30.28 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13331560 114 - 27905053 GCGAGAGAAGAGAAAUGCCGAAGGAUGCGCUGACGCCGGCGCAGUGUCGACUGCUCAGCCGGCGUUGCAGUCGGCGUCGUUCGAUUGUGGGCCACGAGUGGCAACAGUUGCAGU ((.......(((..(((((((....(((...((((((((((((((....)))))...)))))))))))).)))))))..)))((((((..((((....)))).))))))))... ( -54.60) >DroPse_CAF1 99517 108 - 1 GCGAGAGAAGAGAAACACCACAGGCUGC------GUCGGCGCUGCGUUGACUGCGGAGCUGGCAAAGCAGUCGGCGCCGUUCGAUUGGGGGCCACGUGUGGCAACAGUUACAGU .................(((((((((((------((((((.(((((.....))))).))))))...))))))(((.((.........)).)))...)))))............. ( -39.70) >DroSec_CAF1 84007 114 - 1 GCGAGAGAAGAGAAACGCCGAAGGAUGCGCUGACGCCGGCGCAGUGCCGACUGCUCAGCCGGCGCCGCAGUCGGCGUCGUUCGAUUGUGGGCCACGAGUGGCAACAGUUGCAGU ((.......(((..(((((((....((((..(.((((((((((((....)))))...)))))))))))).)))))))..)))((((((..((((....)))).))))))))... ( -55.80) >DroSim_CAF1 88256 114 - 1 GCGAGAGAAGAGAAAUGCCGAAGGAUGCGCUGACGCCGGCGCAGUGCCGACUGCUCAGCCGGCGUCGCAGUCGGCGUCGUUCGAUUGUGGGCCACGAGUGGCAACAGUUGCAGU ((.......(((..(((((((....(((...((((((((((((((....)))))...)))))))))))).)))))))..)))((((((..((((....)))).))))))))... ( -56.20) >DroAna_CAF1 103627 106 - 1 ---ACAAAAGAGUAUC--CUUUG---GGGCUGGCGCUGACGCUGCGUUGACUGCGAAGUCGGCGGUGCUGCUGGCGUCGUUCGAUUGUGGGCCACGUGUGGCAACAGUUGCAGU ---.((((.(.....)--.))))---((((.((((((..(((..((((((((....))))))))..)).)..))))))))))((((((..((((....)))).))))))..... ( -39.20) >DroPer_CAF1 93654 108 - 1 GCGAGAGAAGAGAAACACCAAAGCCUGC------GUCGGCGCUGCGUUGACAGCGGAGCUGGCAGAGCAGUCGGCGCCGUUCGAUUGGGGGCCACGUGUGGCAACAGUUACAGU ......................((((((------((.(((.((..(((((..((((.((((((......)))))).)))))))))..)).)))))))).)))............ ( -37.80) >consensus GCGAGAGAAGAGAAACGCCGAAGGAUGCGCUGACGCCGGCGCAGCGUCGACUGCGCAGCCGGCGGAGCAGUCGGCGUCGUUCGAUUGUGGGCCACGAGUGGCAACAGUUGCAGU ..................................((((((((((......))))...((((((......))))))))))...(((((...((((....))))..)))))))... (-31.69 = -30.28 + -1.41)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:16 2006