
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,311,537 – 13,311,629 |
| Length | 92 |
| Max. P | 0.949181 |

| Location | 13,311,537 – 13,311,629 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -11.98 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13311537 92 + 27905053 U------GUGUGUGUGCCUGUGC-AUUUUUAGUGCUGCUAAUGCAACUCGUUGCAUGGGUGAAUGCGUUGCAUAAAUUGCAAUAUUCAUAAUUAAAUAG .------..((((.(((((((((-(.......(((.......)))......)))))))))).))))((((((.....))))))................ ( -24.52) >DroVir_CAF1 62706 80 + 1 -------------------GUGCAAUUUGUAGGGCCGCUAAUGCAGCACGUUGCGUCCGCAAAUACGUUGCAUAAAUUGCAAUAUUCAUAAUUAAAAGG -------------------.((((((((((.((((.((....)).((.....))))))((((.....)))))))))))))).................. ( -22.30) >DroGri_CAF1 63244 72 + 1 -------------------AUGCAAUUUGCAAUGGCGCUAAUGCAGCACGUUGC--------AUGCGUUGCAUAAAUUGCAAAAUGCAUAAUUAAACAG -------------------(((((.((((((((...((.((((((((.....))--------.))))))))....)))))))).))))).......... ( -25.10) >DroEre_CAF1 65647 94 + 1 ----CGUGUGUGUGUGCCUGUGC-AUUUUUAGUGCUGCUAAUGCAACUCGUUGCACGGGUGAAUGCGUUGCAUAAAUUGCAAUAUUCAUAAUUAAACAG ----..(((((((.(((((((((-(.......(((.......)))......)))))))))).))))((((((.....))))))............))). ( -27.72) >DroYak_CAF1 77096 98 + 1 UGUGCGUGUGUGUGAGCCUGUGC-AUUUUUAGUGCUGCUAAUGCAACUCGUUGCAUGGGUGAAUGCGUUGCAUAAAUUGCAAUAUUCAUAAUUAAACAG ((((.((((((((..((((((((-(.......(((.......)))......)))))))))..))))..((((.....)))))))).))))......... ( -26.92) >DroAna_CAF1 84739 92 + 1 ------UGUGUGCGUGUGUGUGC-AUUUUUAGUGCUGCUAAUGCAGCUCGUUGCAUGGAUAAAUGCGUUGCAUAAAUUGCAAUAUUCAUAAUUAAACAG ------((((.((((((.(((((-(......(.(((((....))))).)..)))))).))..))))((((((.....))))))...))))......... ( -25.90) >consensus _______GUGUGUGUGCCUGUGC_AUUUUUAGUGCUGCUAAUGCAACUCGUUGCAUGGGUGAAUGCGUUGCAUAAAUUGCAAUAUUCAUAAUUAAACAG ....................(((.((((.(((.....)))(((((((.((((.........)))).))))))))))).))).................. (-11.98 = -12.40 + 0.42)

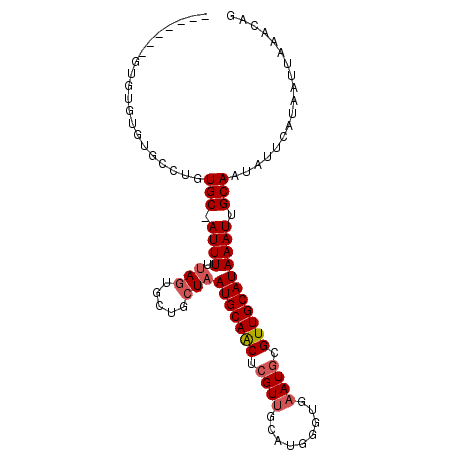

| Location | 13,311,537 – 13,311,629 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -18.49 |
| Consensus MFE | -11.87 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13311537 92 - 27905053 CUAUUUAAUUAUGAAUAUUGCAAUUUAUGCAACGCAUUCACCCAUGCAACGAGUUGCAUUAGCAGCACUAAAAAU-GCACAGGCACACACAC------A .((((((....)))))).(((......((((..((((......)))).....(((((....)))))........)-)))...))).......------. ( -16.00) >DroVir_CAF1 62706 80 - 1 CCUUUUAAUUAUGAAUAUUGCAAUUUAUGCAACGUAUUUGCGGACGCAACGUGCUGCAUUAGCGGCCCUACAAAUUGCAC------------------- ..................((((((((.....((((...(((....)))))))(((((....))))).....)))))))).------------------- ( -19.50) >DroGri_CAF1 63244 72 - 1 CUGUUUAAUUAUGCAUUUUGCAAUUUAUGCAACGCAU--------GCAACGUGCUGCAUUAGCGCCAUUGCAAAUUGCAU------------------- ..........(((((.((((((((...((((.....)--------)))..((((((...)))))).)))))))).)))))------------------- ( -22.10) >DroEre_CAF1 65647 94 - 1 CUGUUUAAUUAUGAAUAUUGCAAUUUAUGCAACGCAUUCACCCGUGCAACGAGUUGCAUUAGCAGCACUAAAAAU-GCACAGGCACACACACACG---- ...........(((((.(((((.....)))))...)))))((.(((((..(.(((((....))))).)......)-)))).))............---- ( -18.60) >DroYak_CAF1 77096 98 - 1 CUGUUUAAUUAUGAAUAUUGCAAUUUAUGCAACGCAUUCACCCAUGCAACGAGUUGCAUUAGCAGCACUAAAAAU-GCACAGGCUCACACACACGCACA ((((......((((((......))))))(((..((((......)))).....(((((....)))))........)-))))))................. ( -16.80) >DroAna_CAF1 84739 92 - 1 CUGUUUAAUUAUGAAUAUUGCAAUUUAUGCAACGCAUUUAUCCAUGCAACGAGCUGCAUUAGCAGCACUAAAAAU-GCACACACACGCACACA------ .((((((....)))))).(((......((((..((((......)))).....(((((....)))))........)-))).......)))....------ ( -17.92) >consensus CUGUUUAAUUAUGAAUAUUGCAAUUUAUGCAACGCAUUCACCCAUGCAACGAGCUGCAUUAGCAGCACUAAAAAU_GCACAGGCACACACAC_______ .................(((((.....))))).((((......)))).....(((((....)))))................................. (-11.87 = -11.90 + 0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:10 2006