
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,304,329 – 13,304,441 |
| Length | 112 |
| Max. P | 0.525513 |

| Location | 13,304,329 – 13,304,441 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -35.11 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13304329 112 + 27905053 CUGCAGCCACCAUUUUGCCACGAUGACAGCUUCAAAUUCACGCACGUCGCAUUUGUGGCAAUUAGUUCGUUGGCAUUCGGAGCUUGAGAGGCUGCG-------AAUGGAAAUCGAUAAU- ...((((.......(((((((((((((.((...........))..))))...))))))))).......)))).((((((.((((.....)))).))-------))))............- ( -32.34) >DroVir_CAF1 57035 114 + 1 CUGCAGCCGCCAUUGUGCCACGAUGACAGCUUUAAAUUUACACAGGUUGCAUUUGUGGCAAUUAGUUCGUUGCCAUUGGGCGCCUGGGAGGCUGUG----UGGAAUGGAAAUUAGU-AU- ..((((((.((...(((((..((((.((((((...........)))))))))).((((((((......))))))))..)))))...)).)))))).----................-..- ( -38.50) >DroGri_CAF1 57093 118 + 1 CUGCAGCCGCCAUUAUGCCACGAUGACAGCUUCAAAUUCACACAGGUUGCAUUUGUGGCAAUUAGUUCAUUGCCAUUGGGCGCCUGCGAGGCUGUGUGUGUGAAAUGGAAAUGAAU-AU- ...((....(((((.((((((....((((((((.........(((((.((....((((((((......))))))))...))))))).))))))))))).))).)))))...))...-..- ( -37.60) >DroWil_CAF1 57516 112 + 1 UCGCAGCCGCCAUUCUGCCACGAUGACAGCUUCAAAUUCACACAUGUCGCAUUGGUGGCAAUUAGUUCAUUACCAUUUGGCGCCUGGGAGGCUGCA--------UUUCGAAUCGAAAUUU ..((((((.((....(((((.((((...................((((((....))))))....((.....)))))))))))....)).))))))(--------(((((...)))))).. ( -32.80) >DroMoj_CAF1 57118 116 + 1 GUGCAGCCACCAUUGUGCCACGAUGACAGCUUCAAAUUCACACAGGUUGCAUUUGUGGCAAUUAGUUCAUUGCCAUUGGGCGCCUGCGAGGCUGUG--CAUGGAAUGUAAAUGAAA-AU- (..(((((..(((((.....)))))...............(.(((((.((....((((((((......))))))))...))))))).).)))))..--).................-..- ( -38.80) >DroAna_CAF1 77640 112 + 1 CCACAGCCGCCGUUGUGCCACGAUGACAACUUCAAAUUCACGCACGUCGCAUUUGUGGCAAUUAGUUCGUUGCCAUUCGGUGCCUGGGAAGCUGGA-------AAUGAAAAUCCAAAAU- ((.(((.(((((.((.((.(((((((.....)))...........(((((....))))).......)))).))))..))))).)))))....((((-------........))))....- ( -30.60) >consensus CUGCAGCCGCCAUUGUGCCACGAUGACAGCUUCAAAUUCACACAGGUCGCAUUUGUGGCAAUUAGUUCAUUGCCAUUGGGCGCCUGGGAGGCUGUG_______AAUGGAAAUCAAA_AU_ ..((((((.(((...((((((((((.(.((...............)).))))).))))))..........((((....))))..)).).))))))......................... (-23.02 = -23.33 + 0.31)

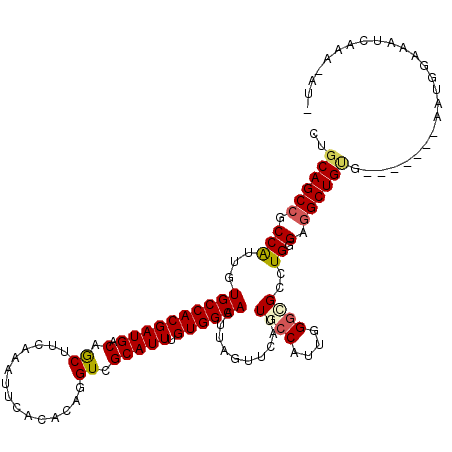

| Location | 13,304,329 – 13,304,441 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -20.51 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13304329 112 - 27905053 -AUUAUCGAUUUCCAUU-------CGCAGCCUCUCAAGCUCCGAAUGCCAACGAACUAAUUGCCACAAAUGCGACGUGCGUGAAUUUGAAGCUGUCAUCGUGGCAAAAUGGUGGCUGCAG -....((((.(((((((-------((.(((.......))).))))))...(((.((...((((.......)))).)).)))))).)))).((.((((((((......)))))))).)).. ( -29.60) >DroVir_CAF1 57035 114 - 1 -AU-ACUAAUUUCCAUUCCA----CACAGCCUCCCAGGCGCCCAAUGGCAACGAACUAAUUGCCACAAAUGCAACCUGUGUAAAUUUAAAGCUGUCAUCGUGGCACAAUGGCGGCUGCAG -..-...............(----(((((((.....)))((....((((((........)))))).....))....)))))........(((((((((.(.....).))))))))).... ( -29.60) >DroGri_CAF1 57093 118 - 1 -AU-AUUCAUUUCCAUUUCACACACACAGCCUCGCAGGCGCCCAAUGGCAAUGAACUAAUUGCCACAAAUGCAACCUGUGUGAAUUUGAAGCUGUCAUCGUGGCAUAAUGGCGGCUGCAG -..-............((((((((....(((.....)))((....(((((((......))))))).....))....))))))))..((.(((((((((..(...)..))))))))).)). ( -36.80) >DroWil_CAF1 57516 112 - 1 AAAUUUCGAUUCGAAA--------UGCAGCCUCCCAGGCGCCAAAUGGUAAUGAACUAAUUGCCACCAAUGCGACAUGUGUGAAUUUGAAGCUGUCAUCGUGGCAGAAUGGCGGCUGCGA ..((((((...)))))--------)((((((..(((..(((((..(((((((......))))))).......((((.((...........))))))....)))).)..))).)))))).. ( -36.70) >DroMoj_CAF1 57118 116 - 1 -AU-UUUCAUUUACAUUCCAUG--CACAGCCUCGCAGGCGCCCAAUGGCAAUGAACUAAUUGCCACAAAUGCAACCUGUGUGAAUUUGAAGCUGUCAUCGUGGCACAAUGGUGGCUGCAC -..-.(((((((((((....((--((..(((.....)))......(((((((......)))))))....))))....)))))))..))))((.((((((((......)))))))).)).. ( -37.90) >DroAna_CAF1 77640 112 - 1 -AUUUUGGAUUUUCAUU-------UCCAGCUUCCCAGGCACCGAAUGGCAACGAACUAAUUGCCACAAAUGCGACGUGCGUGAAUUUGAAGUUGUCAUCGUGGCACAACGGCGGCUGUGG -...(((((........-------)))))...(((((.(.(((..((....)).......((((((..(((((((...((......))..)))).))).))))))...))).).))).)) ( -32.60) >consensus _AU_AUUGAUUUCCAUU_______CACAGCCUCCCAGGCGCCCAAUGGCAACGAACUAAUUGCCACAAAUGCAACCUGUGUGAAUUUGAAGCUGUCAUCGUGGCACAAUGGCGGCUGCAG ..........................(((((.((..(((......((((((........))))))((((((((.....)))..)))))..)))(((.....))).....)).)))))... (-20.51 = -20.23 + -0.27)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:05 2006