
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,303,249 – 13,303,365 |
| Length | 116 |
| Max. P | 0.645603 |

| Location | 13,303,249 – 13,303,365 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -19.84 |
| Energy contribution | -21.12 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13303249 116 + 27905053 GAGCCGG----ACGCACUUACAUCGUUAUGAUGGAAACCGUGAUGAUGACGACGACAAUUAGCGUUGCGAACCAAUCACCGCUGGUCGGAUUUGUUGAGUUUAUCAGAUUGUUCAUCAAA ((.((((----.((......((((((((((..(....))))))))))).((.((((.......))))))..........))))))))((((...((((.....))))...))))...... ( -28.80) >DroVir_CAF1 55925 119 + 1 -AUCCGUCCCCUGGUACUUACAUCGUUAUGAUGGACACUGUGAUUAUGACAACGACAAUAAACGUUGCCAACCAAUCACCGCUGGUUGGAUUUGUCACAUUUAUAAGAUUGUUCAUCAAA -..(((.....)))..............(((((((((.(((((...((((((((((.......))))(((((((........)))))))..))))))...)))))....))))))))).. ( -29.60) >DroGri_CAF1 56067 114 + 1 -AUU-----UCUAGCACUUACAUCGUCAUGAUGGCCACUGUGAUUAUGACAACGACAAUUAACGUUGCCAACCAAUCACCGUUGGUUGGAUUUGUCGCAUUUAUCAGAUUGUUCAUCAAA -...-----....((.........((((((((.((....)).))))))))...(((((.........(((((((((....)))))))))..)))))))...................... ( -29.80) >DroWil_CAF1 56679 108 + 1 ------------GAAACUUACAUCGUUAUGAUGGCCAUUGUGAUUAUGACAAGGACAAUUAAUGUCGCUAACCAAUCACCGUUGGUUGGAUUUGUUGAGUUUAUCAGAUUGUUCAUCAAA ------------................(((((..((.(.((((...((((((((((.....)))).(((((((((....))))))))).))))))......)))).).))..))))).. ( -28.00) >DroMoj_CAF1 55672 112 + 1 -CCCCGUC-------GCUUACAUCGUCAUGAUGGACAGUGUAAUUAUGACAACGACAAUGAACGUUGCCAACCAAUCACCGCUGGUCGGAUUUGUCACAUUUAUCAGAUUGUUCAUCAAA -.......-------.............(((((((((((((((...((((((((((.......))))((.((((........)))).))..))))))...))))...))))))))))).. ( -26.80) >DroAna_CAF1 76777 115 + 1 -AGCAGA----AGGCACUUACAUCGUGAUGAUGGAAACGGUGAUGAUGACAACAACAAUUAGCGUCGAGAACCAAUCACCGCUGGUUGGAUUUGUUGCGUUUAUCAGAUUGUUCAUCAAA -.((...----..))..........((((((((....)).((((((((.(((((((((((((((..((.......))..)))))))))...)))))))).))))))......)))))).. ( -31.60) >consensus _AGCCG______GGCACUUACAUCGUUAUGAUGGACACUGUGAUUAUGACAACGACAAUUAACGUUGCCAACCAAUCACCGCUGGUUGGAUUUGUCGCAUUUAUCAGAUUGUUCAUCAAA ............................(((((((((.(.((((..((((((((((.......))))....(((((((....)))))))..)))))).....)))).).))))))))).. (-19.84 = -21.12 + 1.28)

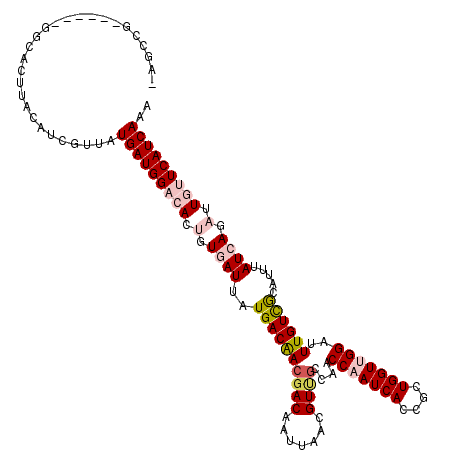

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:02 2006