
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,287,427 – 13,287,532 |
| Length | 105 |
| Max. P | 0.740004 |

| Location | 13,287,427 – 13,287,532 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13287427 105 + 27905053 AGUUG-CU------------UGGCAACAUUACGCCAAUUCAAUUGAUUUGCUCUUCGAGCGCAGUGCGUUUUUAAGCGCAGUGCAAAUAAAACUAAUGCAAUGUGCUCUCUGA-AAAGU (((((-.(------------((((........)))))..))))).....(((.((((((((((.((((((....)))))).((((...........)))).)))))))...))-).))) ( -28.40) >DroPse_CAF1 59300 109 + 1 AGUUG-GCU-------GG-CUGGCAACAUUGCGCCAAUUCAAUUGAUUUGCUUCUGAAGUGCAAUGCGUUUUUGAGCGCUGUGCAAAUAAAACUAAUGCAAUGUGUGCCCCAA-AAACU ((((.-..(-------((-..(((((((((((((.((.((....)).))))........((((..(((((....)))))..))))............))))))).))))))).-.)))) ( -31.10) >DroGri_CAF1 41460 118 + 1 AGUUG-CCUUGGAUGGGGCUUGGCAACAUUGCGCAAAUUCAAUUGAUUUGCCUCUGAAGUGCAGUGCGUUUUUAAGCGCUCCACAAAUACAACUAAUGCAAUGCAUACCCCAAAAAACU .....-.......(((((..((....(((((((((((((.....))))))).......(((.(((((........))))).))).............))))))))..)))))....... ( -33.30) >DroWil_CAF1 43744 106 + 1 AGUUAUCC------------UGGCAACAUUGCGUCAAUUCAAUUGAUUUGCUUUUGAAGUGCAAUGCGUUUUUAAGCGCUGCACAAAUAAAACUAAUGCAAUGUAUGCCCCCA-GAAGU ........------------.(((((((((((((...(((((..(.....)..)))))(((((..(((((....))))))))))...........))))))))).))))....-..... ( -31.10) >DroAna_CAF1 62522 105 + 1 AGUUG-CC------------UGGUAACAUUGCGCCAAUUCAAUUGAUUUGACUUUCGAGUGCAAUGCGUUUUUAAGCGCUGCGCAAACAAAACUAAUGCACUGUGUGCCCUGA-AAGGA .....-((------------((((((((.((((.(((.((....)).)))........(((((..(((((....))))))))))............)))).))).))))....-.))). ( -24.70) >DroPer_CAF1 53286 109 + 1 AGUUG-GCU-------GG-CUGGCAACAUUGCGCCAAUUCAAUUGAUUUGCUUCUGAAGUGCAAUGCGUUUUUGAGCGCUGUGCAAAUAAAACUAAUGCAAUGUGUGCCCCGA-AAACU ((((.-.(.-------((-..(((((((((((((.((.((....)).))))........((((..(((((....)))))..))))............))))))).))))))).-.)))) ( -30.50) >consensus AGUUG_CC____________UGGCAACAUUGCGCCAAUUCAAUUGAUUUGCUUCUGAAGUGCAAUGCGUUUUUAAGCGCUGCGCAAAUAAAACUAAUGCAAUGUGUGCCCCGA_AAACU .....................((((((((((((.(((.((....)).)))........(((((..(((((....))))))))))............)))))))).)))).......... (-20.80 = -21.47 + 0.67)



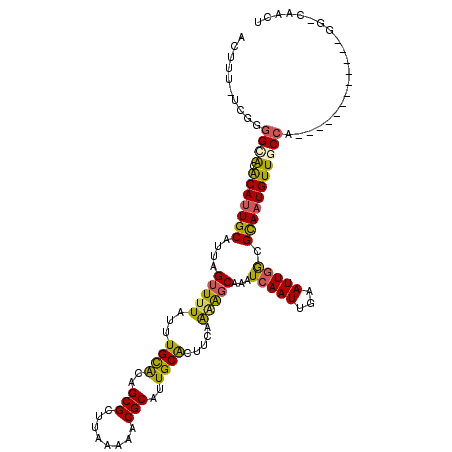
| Location | 13,287,427 – 13,287,532 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -19.94 |
| Energy contribution | -19.28 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13287427 105 - 27905053 ACUUU-UCAGAGAGCACAUUGCAUUAGUUUUAUUUGCACUGCGCUUAAAAACGCACUGCGCUCGAAGAGCAAAUCAAUUGAAUUGGCGUAAUGUUGCCA------------AG-CAACU .....-.....(.(((((((((.(((((((.(((.(((.((((........)))).)))((((...)))).....))).))))))).)))))).)))).------------..-..... ( -26.20) >DroPse_CAF1 59300 109 - 1 AGUUU-UUGGGGCACACAUUGCAUUAGUUUUAUUUGCACAGCGCUCAAAAACGCAUUGCACUUCAGAAGCAAAUCAAUUGAAUUGGCGCAAUGUUGCCAG-CC-------AGC-CAACU ((((.-((((((((.(((((((.(((((((.((((((...(((........))).(((.....)))..)))))).....))))))).)))))))))))..-))-------)).-.)))) ( -28.90) >DroGri_CAF1 41460 118 - 1 AGUUUUUUGGGGUAUGCAUUGCAUUAGUUGUAUUUGUGGAGCGCUUAAAAACGCACUGCACUUCAGAGGCAAAUCAAUUGAAUUUGCGCAAUGUUGCCAAGCCCCAUCCAAGG-CAACU .(((((.((((((..(((((((.............(((.((.((........)).)).))).......(((((((....).)))))))))))))......))))))...))))-).... ( -35.10) >DroWil_CAF1 43744 106 - 1 ACUUC-UGGGGGCAUACAUUGCAUUAGUUUUAUUUGUGCAGCGCUUAAAAACGCAUUGCACUUCAAAAGCAAAUCAAUUGAAUUGACGCAAUGUUGCCA------------GGAUAACU ...((-(...((((.(((((((....(((((....((((((.((........)).))))))....)))))...(((((...))))).))))))))))).------------)))..... ( -32.30) >DroAna_CAF1 62522 105 - 1 UCCUU-UCAGGGCACACAGUGCAUUAGUUUUGUUUGCGCAGCGCUUAAAAACGCAUUGCACUCGAAAGUCAAAUCAAUUGAAUUGGCGCAAUGUUACCA------------GG-CAACU .....-...((..(((...(((.....(((((..((((..(((........)))..))))..)))))(((((.((....)).)))))))).)))..))(------------(.-...)) ( -25.40) >DroPer_CAF1 53286 109 - 1 AGUUU-UCGGGGCACACAUUGCAUUAGUUUUAUUUGCACAGCGCUCAAAAACGCAUUGCACUUCAGAAGCAAAUCAAUUGAAUUGGCGCAAUGUUGCCAG-CC-------AGC-CAACU .....-..((((((.(((((((.(((((((.((((((...(((........))).(((.....)))..)))))).....))))))).)))))))))))..-))-------...-..... ( -27.20) >consensus ACUUU_UCGGGGCACACAUUGCAUUAGUUUUAUUUGCACAGCGCUUAAAAACGCAUUGCACUUCAAAAGCAAAUCAAUUGAAUUGGCGCAAUGUUGCCA____________GG_CAACU ..........((((.(((((((....(((((...((((..(((........)))..)))).....)))))...(((((...))))).)))))))))))..................... (-19.94 = -19.28 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:56 2006