
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,264,547 – 13,264,656 |
| Length | 109 |
| Max. P | 0.637251 |

| Location | 13,264,547 – 13,264,656 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.61 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.53 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13264547 109 - 27905053 GCUCGGGAAAAUCGAACC----AAAUUGGGGGGCCAC-----UG-CCACUUCCUCGAAUGGUGCGACAUGUAAAACGCGUGUCAGGCGACAACGAUGCGUGU-UAACAAUGCGUAAGUAU ((((((.....))))(((----(..(((((((((...-----.)-))....)))))).))))))((((((((.....((((((....))).))).)))))))-)....((((....)))) ( -31.00) >DroVir_CAF1 21863 104 - 1 -CUCG--AAAAGCCCCCU----AAAUAGGG-GG-CAA-----AG-CGACUUCCUCAAAUGGAGCGACAUGUAAAACGCGUGUCAGGUGACAACGAUACAUGU-UAACAAUGCGUAAGUAU -.(((--....(((((((----....))))-))-)..-----..-))).((((......)))).((((((((.....((((((....))).))).)))))))-)....((((....)))) ( -36.20) >DroGri_CAF1 20989 102 - 1 ------AAAUACCCCGCU----AAAAAUGG-GGCUAC-----AG-CGACUUCCUCAAAUGGAGCGACAUGUAAAACGCGUGUCAGGUGACAACGAUACAUGU-UAACAAUGCGUAAGUAU ------.....(((((..----.....)))-))....-----.(-(....(((......)))))((((((((.....((((((....))).))).)))))))-)....((((....)))) ( -25.80) >DroWil_CAF1 22929 116 - 1 --ACAAACAAAUCUGCCC-CA-AAAAUGGGAGGCAGCAACAGCGUCAACUUCCUCGAAUGGGGCGACAUGUAAAACGCGUGUCAUGUGACAUCGAUGCGUGUUUAACAAUGCGUAAGUAU --...........(((((-((-.....(((((...((....)).....))))).....)))))))...(((.((((((((((((((...))).))))))))))).)))((((....)))) ( -39.40) >DroMoj_CAF1 17362 110 - 1 -CUCGCAAAAGGCCCCCUCGAAAAAAGAGG-GG-CAC-----AG-CGACUUCCUCAAAUGGAGCGACAUGUAAAACGCGUGUCAGGUGACAACGAUACAUGU-UAACAAUGCGUAAGUAU -.((((.....(((((.((.......))))-))-)..-----.)-))).((((......)))).((((((((.....((((((....))).))).)))))))-)....((((....)))) ( -35.30) >DroAna_CAF1 41104 106 - 1 -CUCGAAAAAAUCUCUCC----AAAAAUGGGGGCCA-------G-CAACUUCCUCGAAUGGUGCAACAUGUAAAACGCGUGUCAGGCGACAACGAUGCGUGU-UAACAAUGCGUAAGUAU -.(((.........((((----(....)))))(((.-------(-((....((......))))).((((((.....))))))..))).....)))(((((((-.....)))))))..... ( -25.00) >consensus _CUCG_AAAAAUCCCCCC____AAAAAGGG_GGCCAC_____AG_CGACUUCCUCAAAUGGAGCGACAUGUAAAACGCGUGUCAGGUGACAACGAUACAUGU_UAACAAUGCGUAAGUAU .................................................((((......))))..(((((((.....((((((....)))).)).)))))))......((((....)))) (-15.78 = -15.53 + -0.25)

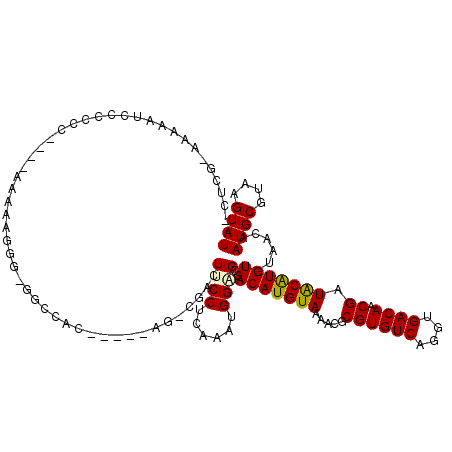
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:48 2006