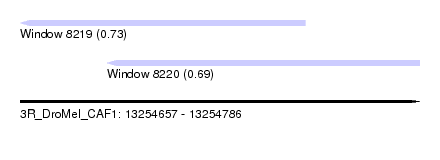
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,254,657 – 13,254,786 |
| Length | 129 |
| Max. P | 0.731843 |

| Location | 13,254,657 – 13,254,749 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.88 |
| Mean single sequence MFE | -22.99 |
| Consensus MFE | -13.95 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731843 |
| Prediction | RNA |
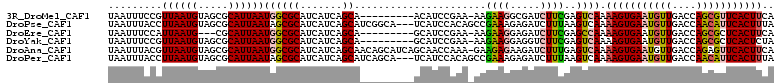
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13254657 92 - 27905053 CAGCA---------ACAUCCGAA-AAGAAGGCGAUCUUCGAGUCAAAAGUGAAUGUUGACCAGCGUUCACUUCAAGAAAUCGAGCAAUUGCAAAAUAUUGU-G ..(((---------((.((....-..)).)(((((.(((((.((..(((((((((((....)))))))))))...))..)))))..)))))......))))-. ( -21.70) >DroPse_CAF1 18448 100 - 1 CAGCAUCGGCA---UCAUCCACAGCCGAAAGAGAUCUUUAAGUCAAAAGUGAAUGUUGACCAACAUUCACUUUAAGAAAUCGAGCAGUUGCAAAAUAUAGGUA ..((((((((.---.........)))))..((..((((.......((((((((((((....))))))))))))))))..)).......)))............ ( -26.21) >DroEre_CAF1 6885 92 - 1 CAGCA---------GCAUCCGAA-AAGAAGGAGAUCUUCGAGCCAAAAGUGAAUGUUGACCAGCGCUCACUUCAAGAAAUCGAGCAGUUGCAAAAUCUUGU-G ..(((---------((.(((...-.....)))....(((((.(...((((((.((((....)))).))))))...)...)))))..)))))..........-. ( -22.80) >DroYak_CAF1 14676 92 - 1 CAGCA---------GCAUCCGAA-AAGAAGGAGGUCUUCGAGUCAAAAGUGAAUGUUGACCAGCGCUCACUCUAAGAAAUCGAGCAGUUGCAAAAUAUUGU-G ..(((---------((.(((...-.....)))((((......(((....))).....))))...((((.............)))).)))))..........-. ( -21.92) >DroAna_CAF1 31178 101 - 1 CAGCAACAGCAUCAGCAACCAAA-GAAGAGAAGAUCUUUGAGUCAAAAGUGAAUGUUGACCAGAGUUCACUUCAAGAAACCGAGCAGUUGCAAAAUAUACU-C ..(((((.((....)).......-((((.(((...(((((.(((((.........))))))))))))).)))).............)))))..........-. ( -22.60) >DroPer_CAF1 19857 100 - 1 CAGCAUCAGCA---UCAUCCACAGCCGAAAGAGAUCUUUAAGUCAAAAGUGAAUGUUGACCAACAUUCACUUUAAGAAAUCGAGCAGUUGCAAAAUAUAGUUA ..(((...((.---((.......((..((((....))))..))..((((((((((((....))))))))))))........))))...)))............ ( -22.70) >consensus CAGCA_________GCAUCCAAA_AAGAAGGAGAUCUUCGAGUCAAAAGUGAAUGUUGACCAGCGUUCACUUCAAGAAAUCGAGCAGUUGCAAAAUAUAGU_G ..(((.....................((((.....)))).......(((((((((((....)))))))))))................)))............ (-13.95 = -13.98 + 0.03)

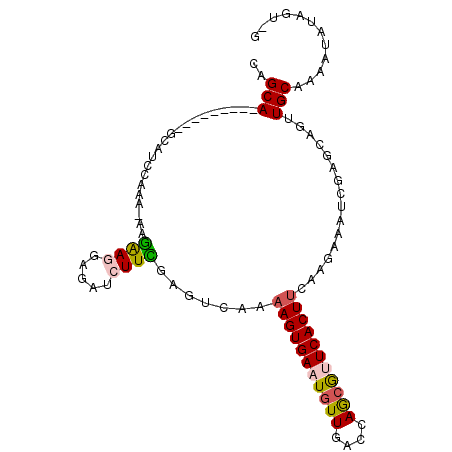
| Location | 13,254,685 – 13,254,786 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13254685 101 - 27905053 UAAUUUCCGUUAAUGUAGCGCAUUAAUGGCGCAUCAUCAGCA---------ACAUCCGAA-AAGAAGGCGAUCUUCGAGUCAAAAGUGAAUGUUGACCAGCGUUCACUUCA ......(((((((((.....))))))))).((.......)).---------......((.-..((((.....))))...))..(((((((((((....))))))))))).. ( -27.40) >DroPse_CAF1 18477 108 - 1 UAAUUUACCUUAAUGUAGCGCAUUAAUAGCGCAUCAUCAGCAUCGGCA---UCAUCCACAGCCGAAAGAGAUCUUUAAGUCAAAAGUGAAUGUUGACCAACAUUCACUUUA ........(((((....((((.......))))..........(((((.---.........))))).........)))))...((((((((((((....)))))))))))). ( -29.50) >DroEre_CAF1 6913 98 - 1 UAAUUUCCAUUAAUG---CGCAUUAAUGGCGCAUCAUCAGCA---------GCAUCCGAA-AAGAAGGAGAUCUUCGAGCCAAAAGUGAAUGUUGACCAGCGCUCACUUCA ..((((((....(((---(((.......))))))........---------...((....-..)).))))))...........((((((.((((....)))).)))))).. ( -20.30) >DroYak_CAF1 14704 101 - 1 UAAUUUCCGUUAAUGUAGCGCAUUAAUGGCGCAUCAUCAGCA---------GCAUCCGAA-AAGAAGGAGGUCUUCGAGUCAAAAGUGAAUGUUGACCAGCGCUCACUCUA .......((((((((.....))))))))((((..........---------...(((...-.....)))((((......(((....))).....)))).))))........ ( -24.90) >DroAna_CAF1 31206 110 - 1 UAAUUUACGUUAAUGUAGCGCAUUAAUGGCGCAUCAUCAGCAACAGCAUCAGCAACCAAA-GAAGAGAAGAUCUUUGAGUCAAAAGUGAAUGUUGACCAGAGUUCACUUCA ........(((.(((..((((.......))))..))).)))....((....)).......-((((.(((...(((((.(((((.........))))))))))))).)))). ( -24.60) >DroPer_CAF1 19886 108 - 1 UAAUUUACCUUAAUGUAGCGCAUUAAUAGCGCAUCAUCAGCAUCAGCA---UCAUCCACAGCCGAAAGAGAUCUUUAAGUCAAAAGUGAAUGUUGACCAACAUUCACUUUA ........((((((((.((((.......)))).......((....)).---......)))..(....)......)))))...((((((((((((....)))))))))))). ( -25.40) >consensus UAAUUUACGUUAAUGUAGCGCAUUAAUGGCGCAUCAUCAGCA_________GCAUCCAAA_AAGAAGGAGAUCUUCGAGUCAAAAGUGAAUGUUGACCAGCGUUCACUUCA .........((((((.....))))))((((((.......))......................((((.....))))..)))).(((((((((((....))))))))))).. (-17.99 = -18.22 + 0.22)
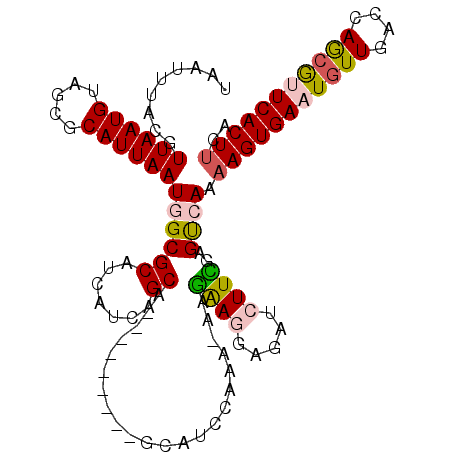
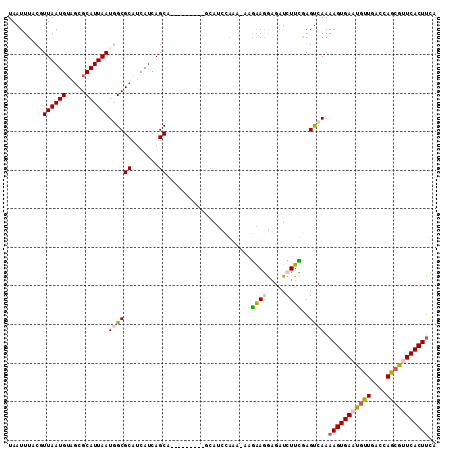
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:42 2006